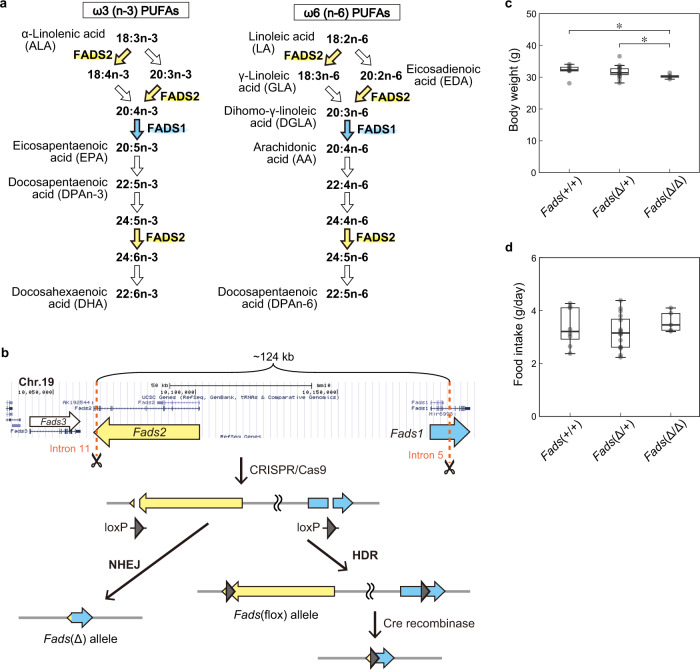
Fig. 1. Generation of mutant mice.
a The PUFA biosynthesis pathways. The desaturation reactions of ω3 and ω6 PUFAs are catalyzed by FADS1 (cyan arrows) or FADS2 (yellow arrows). b A strategy for simultaneous generation of alleles using the CRISPR/Cas9 system. A CRISPR/Cas9 cocktail containing two gRNAs that targeted the Fads1/2 genes and two single-stranded DNA donor templates with the loxP sequence (Supplementary Table 3) was microinjected into mouse fertilized eggs (C57BL/6JJcl strain). Nonhomologous end joining (NHEJ) resulted in the deletion of ~124 kb (Fads(Δ)) and homology-directed repair (HDR) generated the floxed allele. c Body weight of heterozygous and homozygous Fads1/2 deficient mice. Fads(Δ/Δ) mice were significantly leaner than the Fads(Δ/+) and WT (Fads(+/+)) mice (*P < 0.05, d = 0.80 and 1.24 [large effect size (ES)], respectively, t-test with Bonferroni correction). Males, 18–33 weeks old. The box length and a horizontal bar show the interquartile range (IQR) and median, respectively. The length of the whiskers is defined as 1.5 times the upper and lower limits of the IQR. d Food intake. Daily food intake did not differ by genotype.