Figure 5. Assessment of the MXRA8 binding model.
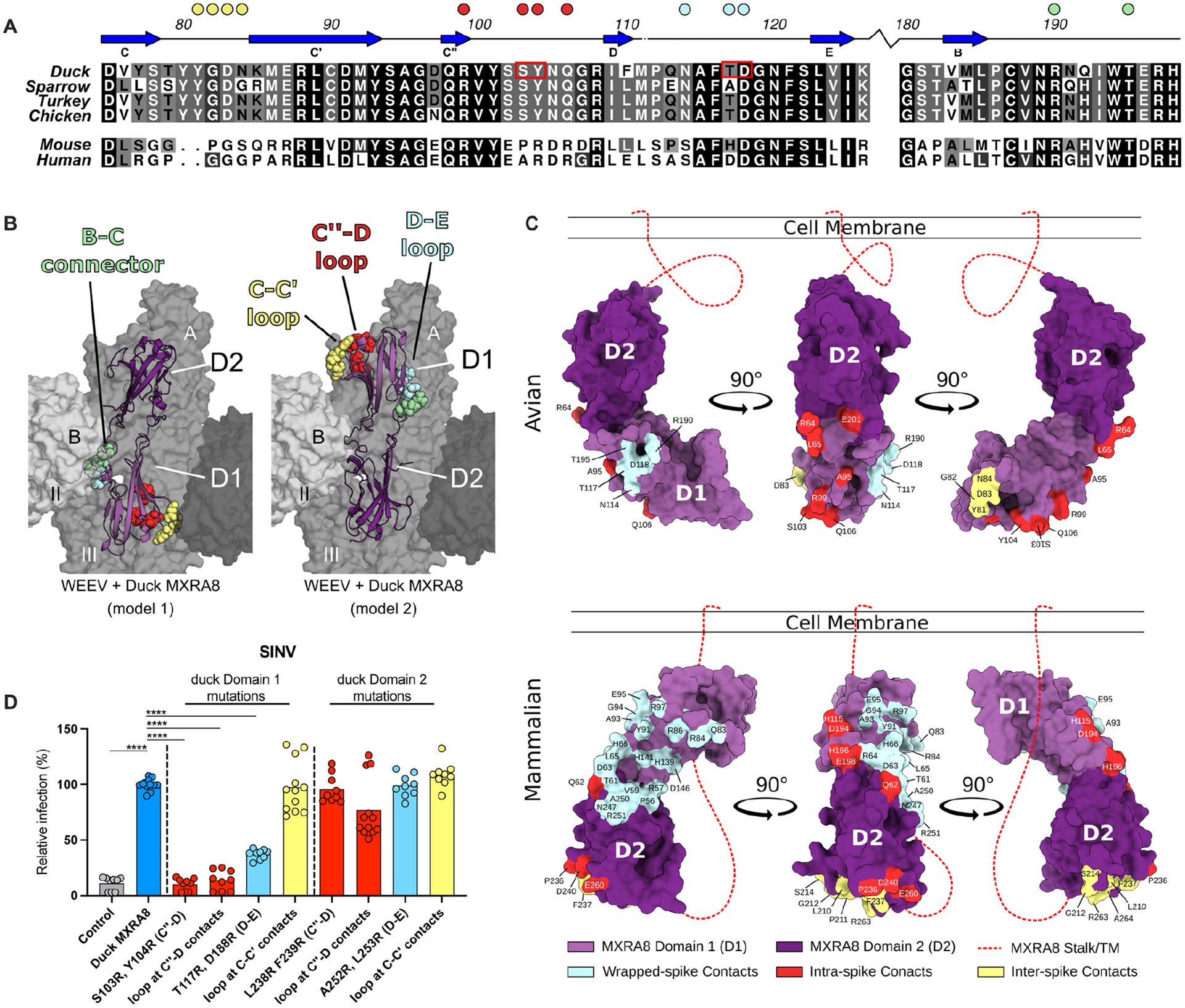
A. Sequence alignment of D1 of MXRA8 at regions that contact WEEV E1. Similar sequence identities are colored by most variable (white) to conserved (black). β-strands are labeled based on topology in Fig 4A. Filled circles above the alignment denote avian MXRA8 contact residues in the C-C’ loop (yellow), C”-D loop (red), D-E loop (cyan), and B-C connector (pale green) are denoted by circles. B. Structure of avian MXRA8 in two possible binding modes, non-flipped (model 1, left) and flipped (model 2, mouse/human-like, right), with contact residues in either D1 or D2 shown as space filling spheres in the C-C’ loop (yellow), C”-D loop (red), D-E loop (cyan), and B-C connector (pale green). C. Binding sites for WEEV and CHIKV on D1 (light purple) and D2 (dark purple) of MXRA8. Structurally defined binding sites and amino acid contact residues are colored according to the viral E2-E1 heterodimer engaged: wrapped (cyan), intraspike (red), or interspike (yellow). Stalk attaching MXRA8 to the cell membrane is represented as a red dashed line and is used to denote mode of binding, with mammalian MXRA8 adopting a flipped orientation relative to avian MXRA8. Avian and mammalian MXRA8 engage E2-E1 heterodimers most proximal to viral membrane with D1 and D2, respectively. D. SINV-GFP infection in ΔMxra8 3T3 cells complemented with wild-type (blue) and indicated mutants (light blue, yellow, or red) duck MXRA8 (3 experiments, triplicate; normalized to wild-type duck MXRA8 infection. Infection and GFP fluorescence were analyzed by flow cytometry. D: one-way ANOVA with Dunnett’s post-test; mean ± standard deviation (SD). ****, P < 0.0001). See also Fig S10.
