Abstract
Human immunodeficiency virus (HIV) or AIDS is currently the leading cause of death in Uganda, with at least three HIV clades (subtypes) accounting for most new infections. Whether an effective vaccine formulated on viruses from a single clade will be able to protect against infection from other local clades remains unresolved. We examined the T-cell immune responses from a cohort of HIV-seropositive individuals in Uganda with predominantly clade A and D infections. Surprisingly, we observed similar frequencies of cross-clade T-cell responses to the gag, env, and nef regions. Our data suggest that the level of viral sequence variability between distinct HIV strains does not predict the degree of cross-clade responses. High sequence homologies were also observed between consensus peptides and sequences from viral isolates, supporting the use of consensus amino acid sequences to identify immunogenic regions in studies of large populations.
Human immunodeficiency virus (HIV) or AIDS continues to be the leading cause of death in sub-Saharan Africa. Although there is a critical need for an effective HIV vaccine, the undefined correlates of protective immunity in HIV type 1 (HIV-1) infection remain an important obstacle. Nevertheless, a wealth of evidence suggests that a robust cellular immune response may curtail viral replication; therefore, HIV vaccine designs have, to date, focused on eliciting strong cellular immune responses.
T-cell epitope recognition is dependent on the individual class I human leukocyte antigen (HLA) alleles. The pattern of antigen recognition and immunodominance may be driven by the prevalence of these alleles in the studied population (19). The identification of highly immunogenic and conserved regions is also complicated by HIV sequence diversity (8). T-cell responses against HIV can, in turn, influence the HIV populations in generating immune escape viral variants (33).
There is comprehensive data on T-cell epitopes for HIV-infected individuals of Caucasian descent. However, there is a relative paucity of information for the geographic regions where the HIV epidemic is spreading the fastest at this time, particularly sub-Saharan Africa (1, 2, 7, 34, 35). In Uganda, clades A and D (as well as A/D recombinant strains) are responsible for approximately 95% of HIV-1 infections (3, 11, 22, 23). Previous studies of HIV-1-specific cellular immune responses in small cohorts of HIV-infected individuals and vaccine recipients in Uganda have shown cross-clade immune recognition (5, 6, 31). Nevertheless, the issue of whether an effective vaccine formulated on viruses from a single clade could protect equally well against viruses from other local clades remains unresolved.
We evaluated the pattern of T-cell antigen recognition in the context of the HLA alleles and multiple circulating HIV subtypes in a cross-sectional study of HIV-infected Ugandan adults. We investigated factors that determine cross-clade T-cell recognition in the context of vaccine design for Uganda and sub-Saharan Africa.
MATERIALS AND METHODS
Study population.
A total of 177 HIV-1-infected adults visiting the HIV clinic at the Joint Clinical Research Centre in Kampala, Uganda, were enrolled in a cross-sectional study. Demographic information was compiled at the time of enrollment and drawing blood. Exclusion criteria included age of <18 years, pregnancy, active tuberculosis, or moribund status. Institutional review board approvals were obtained from the California Department of Health Services and the Joint Clinical Research Center, Kampala, Uganda. All study participants gave written informed consent. The absolute numbers of CD4+ T cells were determined by BD TruCount, and HIV-1 RNA level was determined from plasma using the Roche Amplicor 1.5 (Roche, Branchburg, N.J.) according to the manufacturer's recommendations.
Cell preparation.
Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque (Amersham, Uppsala, Sweden) density centrifugation. Blood was processed within 3 h of drawing blood and setting up for the enzyme-linked immunospot (ELISPOT) assays. Cryopreserved PBMC and plasma were stored in liquid nitrogen and subsequently shipped to the United States for epitope mapping, HLA typing, and viral sequencing.
Antigens.
Peptides corresponding to the sequences of the clade A and D consensus sequences for HIV-1 Gag, Env, and Nef were synthesized as 15-amino-acid (aa) peptides, overlapping by 11 aa (Mitochor Mimotopes, Clayton, Victoria, Australia). Synthetic peptides were pooled for the ELISPOT assay and include three pools of Gag peptides (123 peptides), five pools of Env peptides (210 peptides), and one pool of Nef peptides (49 peptides) for each consensus clade A or D. The final concentration of individual peptides (20 to 40 peptides/pool) was 1 μg/ml per peptide. Consensus sequences were obtained from the Los Alamos database (http://www.hiv.lanl.gov/content/hiv-db/CONSENSUS/M_GROUP/Consensus.html).
Gamma interferon (IFN-γ) ELISPOT assay.
ELISPOT assays were performed on freshly isolated PBMC as previously described (14, 40). Briefly, 96-well nitrocellulose plates (Millititer; Millipore Corp., Bedford, Mass.) were coated with monoclonal antibody 1-D1K (Mabtech, Macka, Sweden) overnight at 4°C. PBMC were added in duplicate wells at 105 cells/well and stimulated with pools of overlapping peptides. All samples were tested for responses against clade A and D gag, env, and nef. Individual peptide responses were confirmed using cryopreserved PBMC, and these responses were confirmed to be CD8-mediated responses by CD8+ T-cell enrichment as previously described (6, 25). Negative and positive controls used were unstimulated (no peptide) and phytohemagglutinin-stimulated (2 μg/ml; Sigma, St. Louis, Mo.) PBMC, respectively. All plates were analyzed using an ELISPOT cytotoxic T lymphocyte (CTL) reader (CTL Analyzers, Cleveland, Ohio). Results were expressed as the number of spot-forming cells (SFC) per 106 PBMC after subtraction of the background value. Positive responses to peptide pools were defined as >100 SFC per 106 PBMC and more than twice the background values. Background values were <10 SFC per 106 PBMC.
HIV sequencing.
Sequencing was performed from frozen plasma samples. RNA was isolated using QIAamp viral RNA isolation kit (QIAGEN, Inc., Valencia, Calif.) according to the manufacturer's instructions. Reverse transcription was performed using the ThermoScript RT system (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions. The Herculase Hotstart DNA polymerase kit (Stratagene, La Jolla, Calif.) was used for PCR. Reverse transcription product was amplified for Nef and Gag using the following specific primers: Nef 5′ primer, CATACCTAGAAGAATAAGACAGGG (RMD8750F); 3′ primer, GCTTATATGCAGCATCTGAGGG (RMD9495R); Gag 5′ primer, GACTAGCGGAGGCTAGAAG (G00 F764); and Gag 3′ primer, AGGGGTCGTTGCCAAAGA (G01 R2264). Sequencing was performed using specific primers including the following primers for Nef: RMD9495R, RMD8750F, TTAAAAGAAAAGGGGGGACTGG (RMD9067F), CCAGTCCCCCCTTTTCTTTTAA (RMD9065R), CTGCCAATCAGGGAAGTAGCCTTGTGT (RMD9143R), and ACACAAGGCTACTTCCCTGATTGGCAG (RMD9143F). Primers for Gag were as follows: G00 F764, G01 R2264, TGTTGGCTCTGGTCTGCTCT (G05 R2138), GTATGGGCAAGCAGGGAGCT (G20 F892), ATTGCYTCAGCCAAAACTCTTGC (G25 R1889), CAGCCAAAATTAYCCTATAGTGCA (G60 F1173), ATTTCTCCYACTGGGATAGGTGG (G55 R1571), ATGCTGARAACATGGGTA (G65 R1312), and TAGAAGAAATGATGACAG (G100 F1817).
Sequencing was performed using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit (ABI, Foster City, Calif.). Cycling conditions were as follows: 3-min incubation at 96°C, followed by 50 cycles, with 1 cycle consisting of 15 s at 96°C, 10 s at 50°C, and 4 min at 60°C. The dyeEx 96 kit procedure (QIAGEN) was used for purification according to the manufacturer's instructions. Combined sequence results were edited using Sequencher V4.1 software (Gene Codes Corporation, Ann Arbor, Mich.), and the sequences were analyzed using ClustalX for alignment and PAUP version 4 (Sinauer Associates, Sunderland, Mass.). The National Institutes of Health's HIV-1 Genotyping Tool (http://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.html) website was used to determine the HIV-1 subtype.
Epitope sequences were aligned and percent homology was determined using the National Center for Biotechnology Information (NCBI) BLAST tool (http://www.ncbi.nlm.nih.gov/BLAST/bl2seq/bl2.html) comparing the patient's viral amino acid sequence to the synthetic consensus peptide sequence. Sequence homology was determined, and the Ugandan Gag and Nef sequences were aligned using the ClustalW tool (http://www.ebi.ac.uk/clustalw/) and PSAweb programs (http://www.imtech.res.in/raghava/psa/) (37). Sequence homologies were analyzed between each index isolate to all other individual isolates for available clade A and D Gag and Nef sequences. Homologies for both intraclade and interclade Gag and Nef sequences were subsequently reported as average percent homologies and standard deviations (SD).
HLA typing.
HLA class I typing was performed by the Stanford Histocompatability Laboratory (Stanford, Calif.) using standard molecular techniques and sequence-specific primers.
Statistical analysis.
Data are presented as arithmetic means ± SD and were compared using the Mann-Whitney test. Linear least-squares regression model was used for statistical analysis in univariate and multivariate analyses. Statistical significance was defined as P values of <0.05.
Nucleotide sequence accession numbers.
Viral nucleotide sequences were submitted to GenBank and given accession numbers AY803354 to AY803488.
RESULTS
Demographic and other characteristics of study participants.
Seventy percent of the 177 enrolled volunteers were antiretroviral (ARV) naïve. The median CD4 cell count was 247 cells/ml (interquartile range, 92 to 332 cells/ml). The median plasma HIV RNA level was 101,120 copies/ml (interquartile range, 16,250 to 359,170 copies/ml). The mean age of the studied cohort was 37.6 ± 7.1 years, and 60.5% (n = 107) were women.
High-resolution HLA class I A and B haplotyping was performed for 44 individuals and included the following distribution in order of frequency: HLA A3001 (4.5%), A0101 (11.4%), B8101 (11.4%), B5301 (11.4%), A0202 (13.6%), B5801 (16%), A7401 (18%), B5802 (27.3%), and A0201 (36%). These results were not dissimilar to the previously reported HLA distribution in the Ugandan population (9).
Subtyping data of Gag and/or Nef open reading frame sequences were available for 62 individuals. Subtyping results using Gag and/or Nef highly concurred with the results of Env subtyping (12, 26, 27, 43). Nef subtyping information was available for 49 samples: 29 samples were subtype A, 19 were subtype D, and 1 was an A/D recombinant. Of the 54 samples with available Gag open reading frame, 28 samples were subtype A, 19 were subtype D, and 7 had recombinant characteristics (4 A/D recombinants, 1 D/C recombinant, 1 A/CRF11 recombinant, and 1 A/G recombinant). Subtyping concurred in 78% of tested individuals between Gag and Nef. No other dominant subtypes were identified from the samples analyzed.
HIV-specific responses by IFN-γ ELISPOT assay.
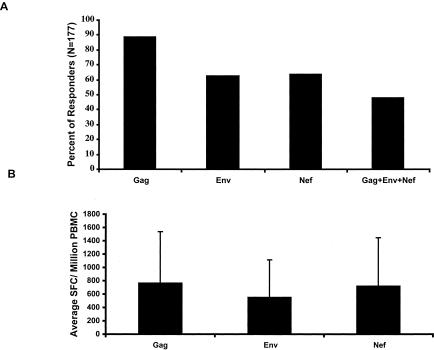
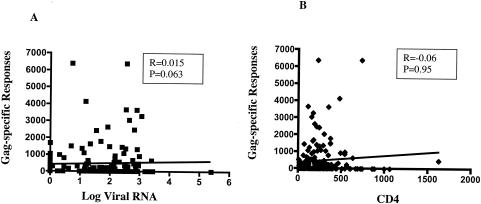
We first examined the HIV-specific responses to Gag, Env, and Nef proteins using the ELISPOT assay. We found that 84.2 and 75.0% of the volunteers demonstrated at least one response against either clade A or D, respectively, with 86 out of 177 volunteers (48.6%) showing responses against all three tested proteins (Fig. 1A). The mean frequencies of responses were highest for Gag, followed by Nef and Env (Fig. 1B). None of the responses against Gag, Env, or Nef correlated with HIV-l load or CD4 cell count, in univariate analyses (Fig. 2; also data not shown). Similar results were also observed in multivariate analyses (controlling for ARV usage and viral load [P > 0.05] [data not shown]).
FIG. 1.
HIV-specific responses against Gag, Env, and Nef as determined by the ELISPOT assays. Freshly isolated PBMC were tested against pooled Gag, Env, or Nef peptides corresponding to clade A and D consensus sequences. (A) Individual responses against Gag, Env, or Nef proteins alone and all three proteins combined for either clade A or D were calculated as the percentage of total volunteers tested (n = 177). (B) Frequencies of responses expressed as mean SFC per 106 PBMC and SD (error bars) were reported for each protein tested.
FIG. 2.
Gag-specific responses as measured by IFN-γ ELISPOT assay do not correlate with clinical disease. No significant correlation (P > 0.05) was observed between the frequency of HIV-specific responses and log HIV RNA level (A) or CD4 cell count (B).
Cross-clade T-cell responses.
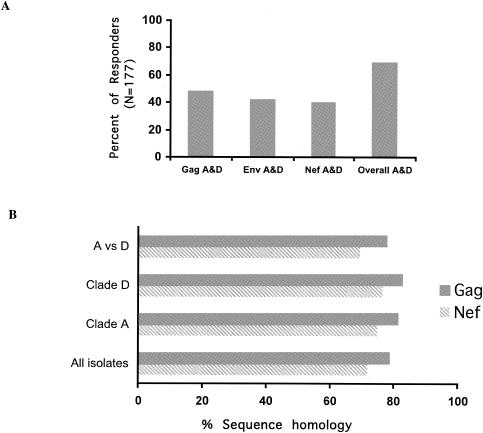
Next we examined the level of cross-clade recognition within each individual tested. Of the 177 volunteers, concordant, positive responses for both clades A and D were observed in 48.6% of responders for Gag (r = 0.61, P < 0.001), 41.8% of responders for Env (r = 0.64, P < 0.001), and 40.1% of responders for Nef (r = 0.58, P < 0.0001) (Fig. 3A). The overall concordance between the two clades for all tested proteins was highly significant (r = 0.69, P < 0.001). Interestingly, no differences in the frequencies of cross-clade responses between clades A and D for Gag, Env, or Nef regions were observed (P > 0.05 [data not shown]), despite the fact that the env region is known to have higher inter- and intraclade sequence variations (16, 17, 41).
FIG. 3.
Cross-clade T-cell responses. (A) Cross-clade recognition against clade A and D (A&D) Gag, Env, and Nef proteins and cross-clade responses against all three tested proteins was calculated as a percentage of individual responders of the total number of volunteers tested (n = 177). (B) Viral sequence identities for clade A and D isolates (within and across clades) were analyzed using ClustalW tool and PSAweb programs, and results are reported as percent homology for either Gag (n = 51) or Nef (n = 44).
Sequence homology across clades has been implicated in the level of cross-clade T-cell recognition. To determine the role of viral subtype on the T-cell response, next we assessed sequence homologies both within and between clades (Fig. 3B). Sequence homologies within clade A for Gag (n = 51) and Nef (n = 44) were 81.4% (SD, 6.33%) and 74.9% (SD, 9.8%), respectively. Similarly, sequence homologies within clade D Gag and Nef were 82.0% (SD, 2.7%) and 73.0% (SD, 8.1%), respectively. Not surprisingly, lower interclade sequence conservation was observed between clade A and D Gag and Nef (77.7% [SD 5.3%] and 69.4% [SD 8.3%], respectively) (Fig. 3B).
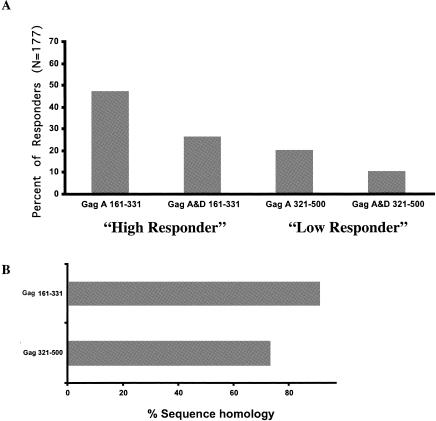
We subsequently assessed whether sequence variability influenced the pattern of T-cell responses. We examined the frequency of responses in different regions of clade A Gag (Fig. 4A) and identified regions of low responders (Gag 321-500 [Gag region from aa 321 to 500], 35 of 177 [19.8%] responders) and epitope-rich or high responders (Gag 161-331, 80 of 177 [46.9%] responders). Stronger intraclade (within clade A) sequence identity in the high-responder compared to the low-responder regions was observed (91.1 versus 73.2%, respectively [P < 0.001] [data not shown]). Cross-clade responses for these Gag regions and sequence homologies to clade D Gag were determined next (Fig. 4). Surprisingly, the dissimilar sequence homologies in the two Gag regions tested did not appear to determine the pattern of cross-clade responses in these regions to clade D. Thus, despite stronger interclade (clade A versus clade D) homology in the high-responder versus low-responder region (89.5 versus 72.9% [P < 0.001] [Fig. 4B]), the same frequency of cross-clade recognition in these two regions was observed (46 cross-recognized clade D antigens out of 83 responders [55.4%] in the high-responder regions versus 18 out of 35 [51.4%] in the low-responder regions [P = 0.737]).
FIG. 4.
Pattern of cross-clade recognition in specific Gag regions. (A) Clade A Gag 321-500 and Gag 161-331 regions were initially designated as low- and high-responder regions, respectively, on the basis of the percentage of positive ELISPOT responses to clade A Gag peptide pools. Cross-clade recognition was subsequently calculated as a percentage of individual clade A responders that cross-recognized clade D peptides corresponding to the respective Gag regions. (B) Specific sequence homologies for the Gag 321-500 and Gag 161-331 regions were calculated as percent homologies of all Gag isolates tested (n = 51).
HIV-specific epitopes.
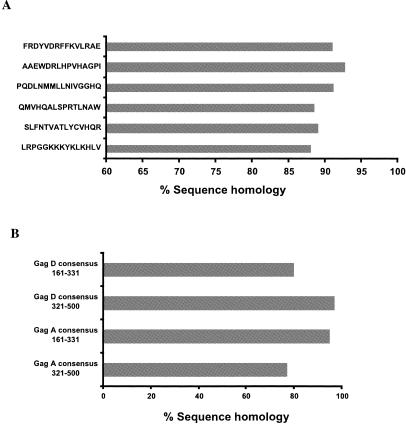
Epitope mapping to individual 15-mer peptides was performed for 10 individual responder samples identified from the pooled peptides by ELISPOT assay (Table 1). All responses were confirmed to be CD8-mediated responses using CD8-enriched T cells as previously described (6, 25). Differences of 0 to 2 aa were observed between the responder 15-mer index peptides and the corresponding autologous sequences. Identified immunogenic epitopes were also compared against reported responses from the Los Alamos T-cell epitope database (http://www.hiv.lanl.gov/content/immunology/tables/ctl_summary.html). We reported several novel T-cell epitopes, characterized by new sequence or HLA restriction. Commonly identified HLA-restricted immunogenic or immunodominant responses were also detected. Interestingly, the highest level of interclade sequence homology was observed for all 15-mer epitopes (Table 1) described in our cohort (range, 86 to 93% [Fig. 5A]), regardless of whether they clustered in conserved or newly identified immunogenic regions.
TABLE 1.
Identified epitopes with 10 associated HLA and sequences
| Peptide sequencea | HLAb | Viral isolate sequencec | Defined restrictiond |
|---|---|---|---|
| LRPGGKKKYRLKHLV | A6601, A6801, B5301, B5802 | LRPGGKKKYKLKHLV | B8 (p17 24-31) |
| -------------I- | A3002, A6801, B5703, B5802 | ||
| SLYNTVATLYCVHQR | A3002, A6801, B5703, B5802 | SLFNTVATLYCVHQR | A3002, A0201, A0202, A0214 B62 (p17 77-85) |
| A0201, A2902, B1402, B1503 | |||
| QMVHQSLSPRTLNAW | A3002, A6801, B5703, B5802 | QMTHQNLSPRTLNAW | B0702, B8101 (p24 16-24) |
| A3001, A6601, B5801, B5802 | QMVHQALSPRTLNAW | ||
| TPQDLNMMLNIVGGH | A0201, A2902, B1402, B1503 | Conserved | B7, B53 (p24 48-56) |
| A0101, A7401, B5801 | |||
| FRDYVDRFYKTLRAE | A0101, A7401, B5801 | FRDYVDRFYKVLRAE | B1801, A2402, B44, B1501, B70 (p24 161-170) |
| --------F------ | |||
| MTYKGAFDLSHFLKE | A6601, A6801, B5301, B5802 | Conserved | No match (Nef 77-85) |
| A0202, A3002, B5703, B5802 | |||
| SKKRQEILDLWVYHT | A0202, A3002, B5703, B5802 | SKKRQDILDLWVYNT | No match (Nef) |
| A6801, A7401, B0702, B3501 | |||
| FPVRPOVPLRPMTYK | A0201, A0301, B4501, B5802 | Conserved | A0301, A11, B35, B51, B27 (Nef 73-82) |
Sequences of responding synthetic peptides. Identified epitopes previously mapped with known HLA restriction are underlined.
HLA A and B typing for individual responders. Underlined haplotypes are previously identified HLA-restricted epitopes.
Autologous sequence matching immunogenic regions with amino acids in bold type representing different amino acids than in the cognate synthetic peptide sequence.
Known published restricted epitopes matching exact sequences of individual synthetic peptides (http://www.hiv.lan1.gov/content/immunology/tables/ctl_summary.html, accessed August 2004). “No match” indicates lack of exact sequence match with known HLA A and B alleles.
FIG. 5.
Pattern of sequence homologies in Gag. (A) High level of homology with viral isolate sequences (clades A and D) is observed for identified epitopes mapped to the 15-mer index peptides. (B) Sequence homologies of consensus sequences corresponding to Gag 321-500 and Gag 161-331 compared to clade A and D viral isolates, respectively.
Use of consensus sequence.
Consensus sequences are commonly used in T-cell assays; however, concerns have been raised that this methodology may skew the pattern of T-cell reactivity. Alignment of clade A and D consensus peptide sequences used in our study demonstrated 81 and 83% homology within Gag clade A and D isolates and 75 and 76% homology within Nef clade A and D isolates, respectively. This homology pattern is not dissimilar from the intraclade sequence homologies within isolates (P > 0.005 [data not shown]). Similarly comparable homologies were also observed between consensus peptides corresponding to the Gag high- and low-responder regions, with 91.9% (SD, 3.2%) and 74.8% (SD, 9.8%) homologies to clade A and 94.5% (SD, 1.9%) and 76.5% (SD, 11.7%) homologies to clade D isolates, respectively (Fig. 5B). The similar homologies between consensus peptides and isolate sequences compared to the homologies within and between clade A and D isolates support our use of these peptides in this study.
DISCUSSION
T-cell-based immunogens remain an important focus in the design of HIV vaccines. Because significant sequence variations exist between and within HIV clades (7 to 15% [29]), disagreement remains as to the importance of clades and their impact on the immune response. We studied an ethnically distinct population in Uganda where multiple HIV strains cocirculate. T-cell immune responses were evaluated irrespective of the infecting clades using the IFN-γ ELISPOT assay and consensus synthetic peptides corresponding to clade A and D HIV proteins. High frequencies of responses were detected in our cohort of predominantly ARV-naïve volunteers. No correlation was found between T-cell responses and clinical disease (as measured by CD4 cell count and HIV RNA level) (15, 35). However, the absence of viral control in the presence of high frequencies of HIV-1-specific T-cell responses as measured by IFN-γ has previously been noted (2, 10, 20). Our data suggest that the effectiveness of virus-specific responses may not be determined solely on the quantitative aspect of IFN-γ responsiveness. Whether specific characteristics of the studied cohort and the infecting clades contribute to the lack of clinical correlation remains unclear.
We found evidence of cross-clade T-cell responses in our studied cohort, similar to previous reports (2a, 6a, 12a, 14a). Interclade sequence variations may be a major determining factor in HLA peptide binding and may abolish T-cell cross-recognition (4, 28, 38). Mutations of the T-cell receptor-interacting amino acids or the HLA-binding amino acids affect T-cell recognition (39); therefore, CD8+ T cells should cross-react poorly across regions with high sequence variability between different infecting clades. Promiscuity of T-cell receptor epitope binding predicted by strong sequence homology has also been suggested (18, 21, 24, 30, 32). The higher interclade Gag and Nef sequence homologies in our cohort would predict more frequent cross-clade T-cell recognition than in the more variable Env region. Surprisingly, we observed comparable levels of cross-clade responses for Gag, Env, and Nef. Thus, cross-clade recognition did not appear to be determined solely by the overall level of sequence homology within distinct HIV regions.
To confirm this observation, we also examined the immune response repertoires in Gag. Within a single clade, we found higher frequencies of responses in conserved regions, whereas areas with higher viral sequence variations have fewer detectable responses. Differential interclade sequence homologies between these respective Gag regions, however, did not appear to affect the respective level of cross-clade recognition. Furthermore, T-cell 15-mer epitopes demonstrated high levels of inter- and intraclade sequence homologies that were not necessarily limited to conserved regions previously described. Thus, immunogenic epitopes even in highly variable proteins or regions combined with distinct HLA profiles may be equally important in determining the pattern and frequency of cross-clade responses within a population.
The phenomenon of epitope clustering has previously been disputed on the basis of a possible bias of the analysis and use of consensus sequences reagents. Consensus sequences may lead to an overestimation in the identification of conserved epitopes (28, 38). Our analysis found similar patterns of sequence homologies between the consensus peptides and individual viral isolates. The lack of observed skewing in the frequency of cross-clade responses to Gag, Env, and Nef lends further support to the use of these reagents in addressing HIV-1 variation in studies of large populations (13, 29, 36, 42).
An effective HIV vaccine remains the likely solution to curbing the AIDS pandemic. The question of whether such a vaccine is applicable for distinct populations and diverse infecting HIV strains persists. One perceived obstacle in the design of a broadly applicable HIV vaccine is the level of viral variability. Our data, however, did not find interclade sequence homologies to be the main determinant of cross-clade T-cell responses. Our study design is primarily focused on the concordance of responses and cannot fully address the effect of sequence homologies on the magnitude and breadth of T-cell responses between clades.
The feasibility of a universal vaccine ultimately rests on the definitive identification of specific T-cell responses and functions that fully correlate with immune protection. Until then, more conclusive evidence on factors determining cross-clade responses will likely require detailed identification of HLA-restricted epitope repertoires in large cohorts to statistically account for both population and viral diversity.
Acknowledgments
This work was supported in part by National Institutes of Health grant AI43885.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Betts, M. R., J. Krowka, C. Santamaria, K. Balsamo, F. Gao, G. Mulundu, C. C. Luo, N. N’Gandu, H. Sheppard, B. Hahn, S. Allen, and J. A. Frelinger. 1997. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J. Virol. 71:8908-8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce, C., J. C. Clegg, A. Featherstone, J. Smith, B. Biryahawaho, R. G. Downing, and J. Oram. 1994. Presence of multiple genetic subtypes of human immunodeficiency virus type 1 proviruses in Uganda. AIDS Res. Hum. Retrovir. 10:1543-1550. [DOI] [PubMed] [Google Scholar]
- 4.Burrows, S. R., S. L. Silins, D. J. Moss, R. Khanna, I. S. Misko, and V. P. Argaet. 1995. T cell receptor repertoire for a viral epitope in humans is diversified by tolerance to a background major histocompatibility complex antigen. J. Exp. Med. 182:1703-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, H., P. Kaleebu, D. Hom, J. Flores, D. Agrawal, N. Jones, J. Serwanga, M. Okello, C. Walker, H. Sheppard, R. El-Habib, M. Klein, E. Mbidde, P. Mugyenyi, B. D. Walker, J. Ellner, R. Mugerwa, and HIV Network for Prevention Trials. 2003. Immunogenicity of a recombinant human immunodeficiency virus (HIV)-canarypox vaccine in HIV-seronegative Ugandan volunteers: results of the HIV Network for Prevention Trials 007 vaccine study. J. Infect. Dis. 187:887-895. [DOI] [PubMed] [Google Scholar]
- 6.Cao, H., I. Mani, R. Vincent, R. Mugerwa, P. Mugyenyi, P. Kanki, J. Ellner, and B. D. Walker. 2000. Cellular immunity to HIV-1 clades: relevance to HIV-1 vaccine trials in Uganda. J. Infect. Dis. 182:1350-1356. [DOI] [PubMed] [Google Scholar]
- 6a.Cao, H., P. Kanki, J.-L. Sankale, A. Dieng-Sarr, G. P. Mazzara, S. A. Kalams, B. Korber, S. MBoup, and B. D. Walker. 1997. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J. Virol. 71:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, J., J. McNevin, S. Holte, L. Fink, L. Corey, and M. J. McElrath. 2003. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J. Virol. 77:6867-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, K., J. Hollenbach, X. Shi, W. Shi, M. Chopek, and M. A. Fernandez-Vina. 2001. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62:1009-1030. [DOI] [PubMed] [Google Scholar]
- 9.Cao, K., A. M. Moormann, K. E. Lyke, C. Masaberg, O. P. Sumba, O. K. Doumbo, D. Koech, A. Lancaster, M. Nelson, D. Meyer, R. Single, R. J. Hartzman, C. V. Plowe, J. Kazura, D. L. Mann, M. B. Sztein, G. Thomson, and M. A. Fernandez-Vina. 2004. Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens 63:293-325. [DOI] [PubMed] [Google Scholar]
- 10.Champagne, P., G. S. Ogg, A. S. King, C. Knabenhans, K. Ellefsen, M. Nobile, V. Appay, G. P. Rizzardi, S. Fleury, M. Lipp, R. Forster, S. Rowland-Jones, R. P. Sekaly, A. J. McMichael, and G. Pantaleo. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410:106-111. [DOI] [PubMed] [Google Scholar]
- 11.Collins, K. R., M. E. Quinones-Mateu, M. Wu, H. Luzze, J. L. Johnson, C. Hirsch, Z. Toosi, and E. J. Arts. 2002. Human immunodeficiency virus type 1 (HIV-1) quasispecies at the sites of Mycobacterium tuberculosis infection contribute to systemic HIV-1 heterogeneity. J. Virol. 76:1697-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelissen, M., G. Kampinga, F. Zorgdrager, J. Goudsmit, and the UNAIDS Network for HIV Isolation and Characterization. 1996. Human immunodeficiency virus type 1 subtypes defined by env show high frequency of recombinant gag genes. J. Virol. 70:8209-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Durali, D., J. Morvan, F. Letourneru, D. Schmitt, N. Guegan, M. Dalod, S. Saragosti, D. Sicard, J.-P. Levy, and E. Gomard. 1998. Cross-reactions between the cytotoxic T-lymphocyte responses of human immunodeficiency virus-infected African and European patients. J. Virol. 72:3547-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellenberger, D. L., B. Li, L. D. Lupo, S. M. Owen, J. Nkengasong, M. S. Kadio-Morokro, J. Smith, H. Robinson, M. Ackers, A. Greenberg, T. Folks, and S. Butera. 2002. Generation of a consensus sequence from prevalent and incident HIV-1 infections in West Africa to guide AIDS vaccine development. Virology 302:155-163. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari, G., J. R. Currier, M. E. Harris, S. Finkelstein, A. de Oliveira, D. Barkhan, J. H. Cox, M. Zeira, K. J. Weinhold, N. Reinsmoen, F. McCutchan, D. L. Birx, S. Osmanov, and S. Maayan. 2004. HLA-A and -B allele expression and ability to develop anti-Gag cross-clade responses in subtype C HIV-1-infected Ethiopians. Hum. Immunol. 65:648-659. [DOI] [PubMed] [Google Scholar]
- 14a.Ferrari, G., W. Humphrey, M. J. McElrath, J. Excler, A. Duliege, M. L. Clements, L. C. Corey, D. P. Bolognesi, and K. J. Weinhold. 1997. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc. Natl. Acad. Sci. USA 94:1396-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. StJohn, A. Khatri, K. Davis, J. Mullins, P. J. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, F., L. Yue, D. L. Robertson, S. C. Hill, H. Hui, R. J. Biggar, A. E. Neequaye, T. M. Whelan, D. D. Ho, and G. M. Shaw. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433-7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavin, M. A., B. Dere, A. G. Grandea III, K. A. Hogquist, and M. J. Bevan. 1994. Major histocompatibility complex class I allele-specific peptide libraries: identification of peptides that mimic an H-Y T cell epitope. Eur. J. Immunol. 24:2124-2133. [DOI] [PubMed] [Google Scholar]
- 19.Goulder, P. J., C. Brander, K. Annamalai, N. Mngqundaniso, U. Govender, Y. Tang, S. He, K. E. Hartman, C. A. O'Callaghan, G. S. Ogg, M. A. Altfeld, E. S. Rosenberg, H. Cao, S. A. Kalams, M. Hammond, M. Bunce, S. I. Pelton, S. A. Burchett, K. McIntosh, H. M. Coovadia, and B. D. Walker. 2000. Differential narrow focusing of immunodominant human immunodeficiency virus Gag-specific cytotoxic T-lymphocyte responses in infected African and Caucasoid adults and children. J. Virol. 74:5679-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harari, A., S. Petitpierre, F. Vallelian, and G. Pantaleo. 2004. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood 103:966-972. [DOI] [PubMed] [Google Scholar]
- 21.Hemmer, B., M. Jacobsen, and N. Sommer. 2000. Degeneracy in T-cell antigen recognition—implications for the pathogenesis of autoimmune diseases. J. Neuroimmunol. 107:148-153. [DOI] [PubMed] [Google Scholar]
- 22.Hoelscher, M., W. E. Dowling, E. Sanders-Buell, J. K. Carr, M. E. Harris, A. Thomschke, M. L. Robb, D. L. Birx, and F. McCutchan. 2002. Detection of HIV-1 subtypes, recombinants, and dual infections in east Africa by a multi-region hybridization assay. AIDS 16:2055-2064. [DOI] [PubMed] [Google Scholar]
- 23.Hu, D. J., T. J. Dondero, M. A. Rayfield, R. George, G. Schochetman, H. W. Jaffe, C. C. Luo, M. L. Kalish, B. G. Weniger, C. Pau, C. A. Schable, and J. W. Curran. 1996. The emerging genetic diversity of HIV. The importance of global surveillance for diagnostics, research and prevention. JAMA 275:210-216. [PubMed] [Google Scholar]
- 24.Jacobsen, M., S. Cepok, W. H. Oertel, N. Sommer, and B. Hemmer. 2001. New approaches to dissect degeneracy and specificity in T cell antigen recognition. J. Mol. Med. 79:358-367. [DOI] [PubMed] [Google Scholar]
- 25.Jones, N., D. Agrawal, M. Elrefaei, A. Hanson, V. Novitsky, J. T. Wong, and H. Cao. 2003. Evaluation of antigen-specific responses using in vitro enriched T cells. J. Immunol. Methods 274:139-147. [DOI] [PubMed] [Google Scholar]
- 26.Jubier-Maurin, V., S. Saragosti, J. L. Perret, E. Mpoudi, E. Esu-Williams, C. Mulanga, F. Liegeois, M. Ekwalanga, E. Delaporte, and M. Peeters. 1999. Genetic characterization of the nef gene from human immunodeficiency virus type 1 group M strains representing genetic subtypes A, B, C, E, F, G, and H. AIDS Res. Hum. Retrovir. 15:23-32. [DOI] [PubMed] [Google Scholar]
- 27.Kaleebu, P., J. Whitworth, L. Hamilton, A. Rutebemberwa, F. Lyagoba, D. Morgan, M. Duffield, B. Biryahwaho, B. Magambo, and J. Oram. 2000. Molecular epidemiology of HIV type 1 in a rural community in southwest Uganda. AIDS Res. Hum. Retrovir. 16:393-401. [DOI] [PubMed] [Google Scholar]
- 28.Klenerman, P., S. Rowland-Jones, S. McAdam, J. Edwards, S. Daenke, D. Lalloo, B. Koppe, W. Rosenberg, D. Boyd, A. Edwards, et al. 1994. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature 369:403-407. [DOI] [PubMed] [Google Scholar]
- 29.Korber, B., B. Gaschen, K. Yusim, R. Thakallapally, C. Kesmir, and V. Detours. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19-42. [DOI] [PubMed] [Google Scholar]
- 30.Mason, D. 1998. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19:395-404. [DOI] [PubMed] [Google Scholar]
- 31.McAdams, S., P. Kaleebu, P. Krausa, P. Goulder, N. French, B. Collin, T. Blanchard, J. Whitworth, A. McMichael, and F. Gotch. 1998. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. AIDS 12:571-579. [DOI] [PubMed] [Google Scholar]
- 32.McKinney, D. M., R. Skvoretz, B. D. Livingston, C. C. Wilson, M. Anders, R. W. Chesnut, A. Sette, M. Essex, V. Novitsky, and M. J. Newman. 2004. Recognition of variant HIV-1 epitopes from diverse viral subtypes by vaccine-induced CTL. J. Immunol. 173:1941-1950. [DOI] [PubMed] [Google Scholar]
- 33.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 34.Novitsky, V., H. Cao, N. Rybak, P. Gilbert, M. F. McLane, S. Gaolekwe, T. Peter, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2002. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J. Virol. 76:10155-10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novitsky, V., U. R. Smith, P. Gilbert, M. F. McLane, P. Chigwedere, C. Williamson, T. Ndung'u, I. Klein, S. Y. Chang, T. Peter, I. Thior, B. T. Foley, S. Gaolekwe, N. Rybak, S. Gaseitsiwe, F. Vannberg, R. Marlink, T. H. Lee, and M. Essex. 2002. Human immunodeficiency virus type 1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J. Virol. 76:5435-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raghava, G. P. 2001. A graphical web server for the analysis of protein sequences and alignment. Biotech Software Internet Rep. 2:225-228. [Google Scholar]
- 38.Reid, S. W., S. McAdam, K. J. Smith, P. Klenerman, C. A. O'Callaghan, K. Harlos, B. K. Jakobsen, A. J. McMichael, J. I. Bell, D. I. Stuart, and E. Y. Jones. 1996. Antagonist HIV-1 Gag peptides induce structural changes in HLA B8. J. Exp. Med. 184:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rentenaar, R. J., L. E. Gamadia, N. van der Hoek, F. N. van Diepen, P. M. Wertheim-van Dillen, J. F. Weel, S. Surachno, R. A. van Lier, and I. J. ten Berge. 2001. CD8(POS) lymphocyte dynamics in primary CMV infection. Transplant. Proc. 33:1867-1869. [DOI] [PubMed] [Google Scholar]
- 40.Russell, N. D., M. G. Hudgens, R. Ha, C. Havenar-Daughton, and M. J. McElrath. 2003. Moving to human immunodeficiency virus type 1 vaccine efficacy trials: defining T cell responses as potential correlates of immunity. J. Infect. Dis. 187:226-242. [DOI] [PubMed] [Google Scholar]
- 41.Spira, S., M. A. Wainberg, H. Loemba, D. Turner, and B. G. Brenner. 2003. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J. Antimicrob. Chemother. 51:229-240. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, C. C., D. McKinney, M. Anders, S. MaWhinney, J. Forster, C. Crimi, S. Southwood, A. Sette, R. Chesnut, M. J. Newman, and B. D. Livingston. 2003. Development of a DNA vaccine designed to induce cytotoxic T lymphocyte responses to multiple conserved epitopes in HIV-1. J. Immunol. 171:5611-5623. [DOI] [PubMed] [Google Scholar]
- 43.Yirrell, D. L., P. Kaleebu, D. Morgan, C. Watera, B. Magambo, F. Lyagoba, and J. Whitworth. 2002. Inter- and intragenic intersubtype HIV-1 recombination in rural and semiurban Uganda. AIDS 16:279-286. [DOI] [PubMed] [Google Scholar]