Figure 8.
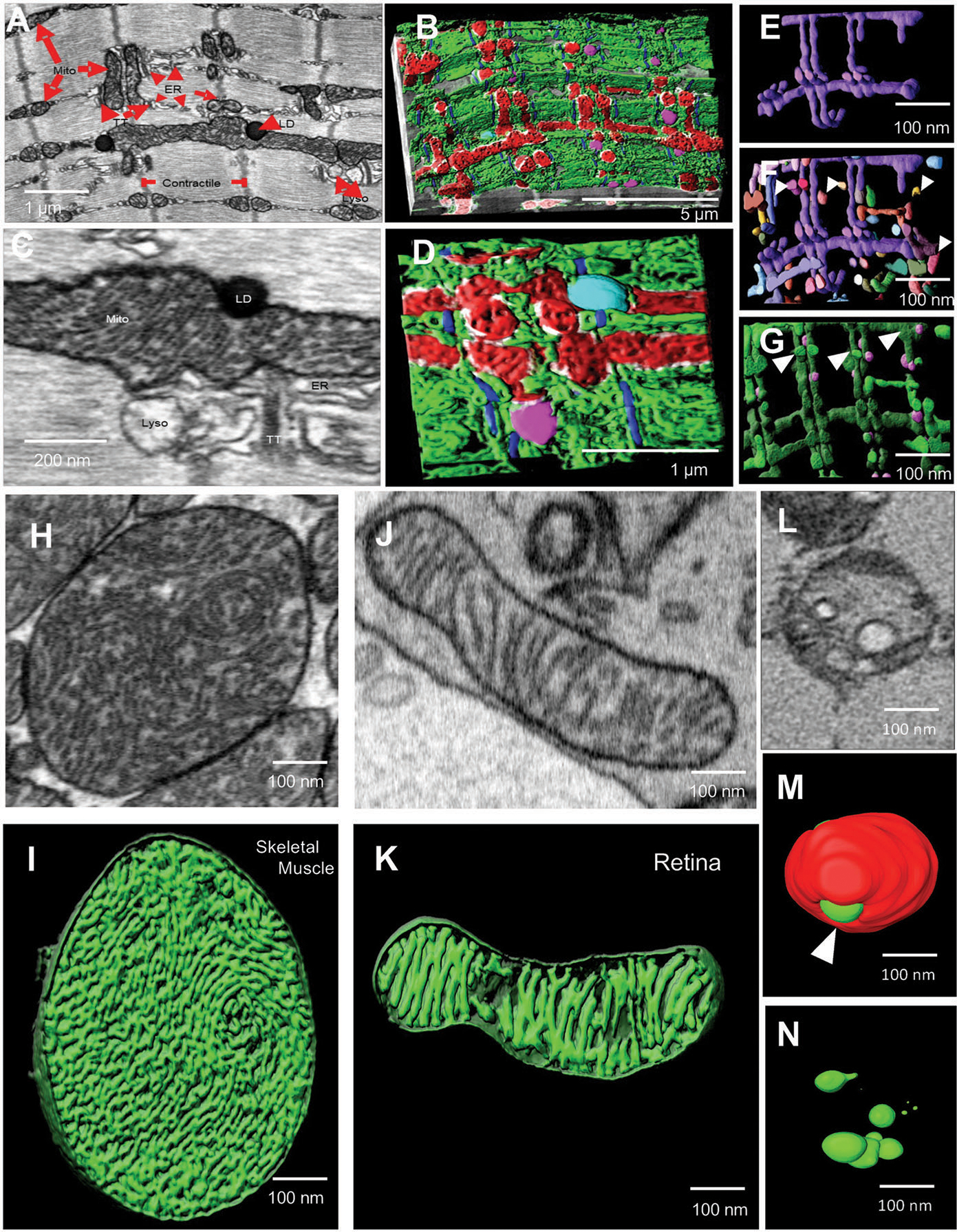
3D reconstruction using FIB-SEM allows identification of complex cellular interactions with high resolution. A–D) TEM micrograph (A) and 3D reconstruction using FIB-SEM (B) of MERCs in mouse skeletal muscle. C) TEM micrograph and D) 3D reconstruction using FIB-SEM of mitochondria (Mito, red), actin (blue), lysosomes (Lyso, pink), lipid droplets (LD, teal), MERCs (white), and ER (green). E–G) 3D reconstruction using FIB-SEM allows the visualization of a single hyperbranched mitochondrion (purple) (E), surrounded by nearby mitochondria (F, various colors, white arrowheads) and lysosomes (G, pink) from adult mouse skeletal muscle. H) TEM micrograph of cristae-containing mitochondria from WT mouse skeletal muscle. I) 3D reconstruction of cristae morphology in WT mouse skeletal muscle using FIB-SEM. J) Micrograph of cristae-containing mitochondria from WT mouse retina tissue. K) 3D reconstruction of cristae morphology in WT mouse retina tissue using FIB-SEM. L) TEM micrograph of a mitochondrion in an Opa1-like KD Drosophila flight muscle. M) 3D reconstruction of cristae (green) with mitochondria (red) in Opa1-like KD Drosophila flight muscle. N) 3D reconstruction of cristae morphology (green) in Opa1-like KD Drosophila flight muscle using SBF-SEM.
