FIG. 1.
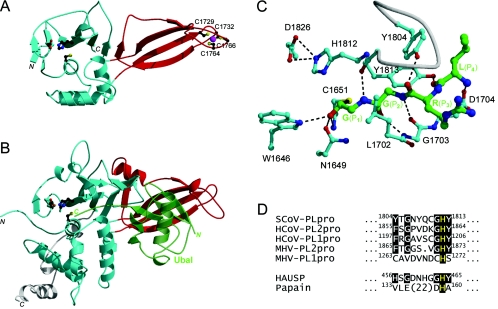
Structure-to-function relationships between SCoV PLpro and HAUSP. (A) Three-dimensional model of SCoV PLpro. The protease domain is rendered in cyan, and the circularly permuted Zn ribbon domain is shown in red. The zinc ion is shown as a magenta sphere, and the Zn-chelating cysteines and the catalytic triad are shown in ball-and-stick configurations. The initial homology model was built as a chimera of two templates, the structure of HAUSP complexed with ubiquitin aldehyde (Ubal) (PDB code, 1NBF), with parts of the protease domain outside the binding site based on the structure of the foot-and-mouth disease virus leader protease (PDB code, 1QMY). The model was refined by gradual structural relaxation by using stepwise energy minimization with the AMBER force field. The Ubal C-terminal sequence RLRG-Glycinal, covalently bound at the catalytic C1651 residue as a hemiacetal adduct, was included during the initial stages of structural refinement and replaced with full-length Ubal in the final stage. (B) Crystal structure of the catalytic core domain of HAUSP with bound Ubal. Colors are as described for panel A, except for those for Ubal (semitransparent green) and the C-terminal extension outside the papain-like fold (white). (C) Specific substrate recognition in S1-through-S4 binding site region of SCoV PLpro. The Ubal C-terminal sequence LRG-Glycinal, also corresponding to the SCoV PLpro cleavage motif, is covalently bound at the catalytic C1651 residue, which together with H1812 and D1826 forms the putative catalytic triad. W1646 serves as the putative oxyanion hole residue. The carbon atoms are colored in green for the ligand and in cyan for the binding site. A binding site loop covering the substrate is depicted as a white tube. Hydrogen bonds are shown with dashed lines. (D) Alignment of the S2 subsite signature sequences from selected coronaviral PLpros, HAUSP, and papain. Conserved residues are highlighted on a black background, with the catalytic histidines being shown in yellow. The predicted critical determinants of the S2 subsite specificity correspond to positions 1804 and 1813 of SCoV PLpro.