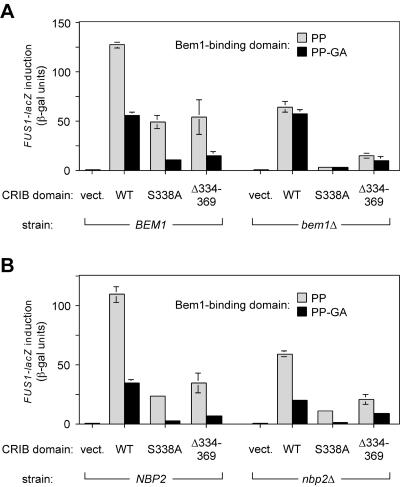
FIG. 5.
The phenotypic effect of Ste20 PxxP mutation depends on Bem1 rather than on Nbp2. The level of FUS1-lacZ induced by α-factor was assayed for GFP-Ste20 fusions that contained combinations of mutations in two domains—the indicated CRIB domain variant (wild type [WT], S338A, or Δ334-369) combined with a Bem1-binding domain that was either intact (PP) or mutant (PP-GA). Control transformants harbored the pRS316 vector (vect.). Deletion mutant strains were compared to congenic wild-type cells in the same strain background. Because both bem1Δ and nbp2Δ mutations impair growth with severities that depend on the strain background, the experiments for which results are shown here were performed in strain backgrounds where the mutants are the least growth impaired (15Dau for BEM1 versus bem1Δ; W303 for NBP2 versus nbp2Δ), but similar results were obtained for nbp2Δ mutants in the strain background (15Dau; PPY1343 versus PPY1415) where the deletion caused a stronger growth defect (data not shown). (A) Deletion of BEM1 mimics the effect of the PP-GA mutation, and in bem1Δ cells the PP-GA mutation confers no further pheromone response defect, indicating that the effect of the PP-GA mutation can be attributed to a disruption of Bem1-Ste20 interaction. Strains: PPY1343 (BEM1) and PPY1341 (bem1Δ). (B) In nbp2Δ cells, the PP-GA mutation still confers a signaling defect. Strains: PPY913 (NBP2) and PPY1456 (nbp2Δ).