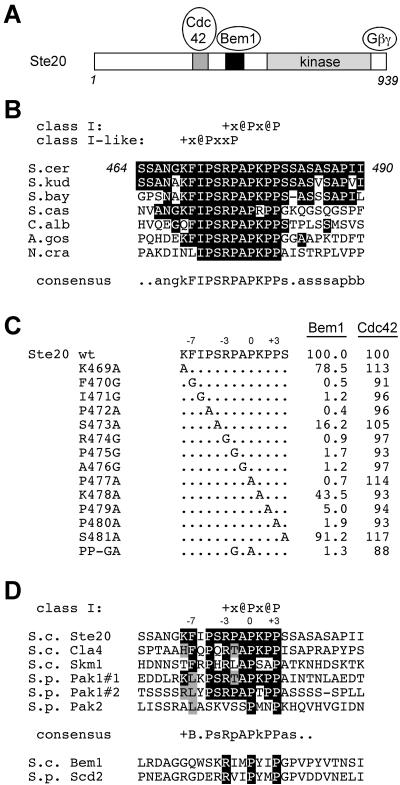
FIG. 7.
Evolutionary conservation of the Bem1 binding site in Ste20 orthologs and paralogs. Standard single-letter symbols for amino acid residues are used. In addition, the symbols +, @, b, and x stand for basic, aliphatic, hydrophobic, and any residues, respectively. (A) Schematic diagram indicating relative positions of defined domains in Ste20. (B) Alignment of sequences from Ste20 orthologs from fungi of increasing phylogenetic distance. Sequences are from S. cerevisiae (S.cer), Saccharomyces kudriavzevii (S.kud), Saccharomyces bayanus (S.bay), Saccharomyces castelli (S.cas), Candida albicans (C.alb), Ashbya gossypii (A.gos), and Neurospora crassa (N.cra). Each sequence shown lies between the CRIB and kinase domains. Identities to the Ste20 sequence are boxed black. (C) Bem1 binding is disrupted by mutation of conserved residues in the Ste20 motif. The indicated point mutations (from K469A to S481A) were incorporated into a DBD-Ste201-499 fusion construct (pB20N2), and interaction with Bem1157-551 (pRL51.1) or Cdc42 (pPP1027) was tested in the two-hybrid tester strain PPY760 by a quantitative β-galactosidase assay. For ease of comparison, signals were normalized to those of wild-type (wt) pB20N2 (taken as 100%), whose mean values were 15.0 (Bem1) and 271.5 (Cdc42) β-galactosidase units; standard deviations (n = 3)were within 15% of the mean in all cases and are omitted for clarity. Note that the mutations do not disrupt interaction with Cdc42 and thus are specific for Bem1. Also, similar Bem1-binding results were observed by using a full-length Bem1 (residues 1 to 551) construct (pXP-BEM1), and no appreciable interaction signals were observed by using a vector (pGAD424) control (data not shown). (D) Alignment of Ste20 paralogs and orthologs from S. cerevisiae (S.c.) and Schizosaccharomyces pombe (S.p.). A variable number of PxxP motifs (from 1 to 12) are present in each protein. Shown here are the only motifs that are perfect matches to class I (+x@Px@P) or class II (@Px@Px+) consensus sequences (45). Exceptions are as follows: Pak2 has no perfect matches, so its only PxxP motif is shown for comparison; Ste20 has one class I match and one class II match, but only the class I match is shown because it is the one identified in this study as a Bem1-binding site. Pak1 has two class I matches (#1 and #2), each of which is shown. Within the core motif, identities to Ste20 are boxed black, and conservative or common substitutions are boxed grey. The top six sequences are from PAK family kinases and thus represent candidate trans ligands for the SH3-2 domain of Bem1 or its ortholog (with the exception of the Pak2 sequence, which is a poor match and which lies within the CRIB domain). The bottom two sequences lie within the PX domains of Bem1 and its S. pombe ortholog, Scd2, and have been proposed to bind in cis to SH3-2 domains within the same protein (21). These sequences match the class I consensus but lack the additional conserved sequence features common to the PAK motifs.