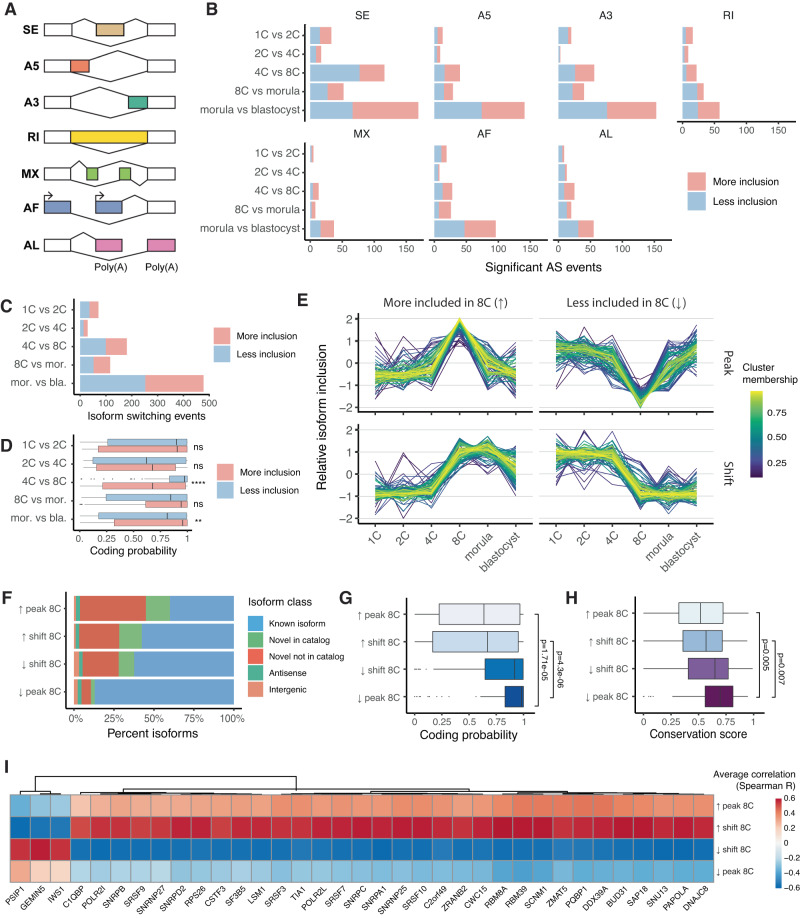
Fig. 5. Alternative splicing induces ORF disruption and novel isoform inclusion during embryonic genome activation.
A Schematic representation of the seven types of AS events analyzed: skipped exon (SE), alternative 5’ splice site (A5), alternative 3’ splice site (A3), retained intron (RI), mutually exclusive exons (MX), alternative first exon (AF), and alternative last exon (AL). B Number of significant AS events at each developmental stage transition. C Number of significant isoform switching events at each developmental stage transition. D Predicted protein-coding probability of isoforms undergoing significant isoform switches at each developmental stage transition, grouped by direction of inclusion. (1C vs 2C, p = 0.94; 2C vs 4C, p = 0.83; 4C vs 8C, p = 4.3e−06; 8C vs morula, p = 0.28; morula vs blastocyst, p = 0.0021, unpaired two-sided Wilcoxon Rank Sum test, Benjamini-Hochberg correction; for 1C vs 2C, n = 35 more included and n = 36 less included isoforms; for 2C vs 4C, n = 16 more included and n = 13 less included isoforms; for 4C vs 8C, n = 82 more included and n = 99 less included isoforms; for 8C vs morula, n = 64 more included and n = 52 less included isoforms; for morula vs blastocyst, n = 226 more included and n = 252 less included isoforms). E Relative inclusion levels of isoforms undergoing significant switching events at the 8C stage. Four major clusters are identified, based on the direction of inclusion (more or less included at the 8C stage) and temporal inclusion dynamics (transient peak or sustained shift). F Structural classes of the isoform clusters defined in E. G, H Coding probabilities and evolutionary conservation scores for the isoform clusters defined in E. (unpaired two-sided Wilcoxon Rank Sum test, Benjamini-Hochberg correction; cluster sizes: n = 80 for positive peak at 8C; n = 92 for positive shift at 8C, n = 93 for negative shift at 8C, n = 85 for negative peak at 8C. Box plot colors are proportional to median values for each corresponding box). I Heatmap displaying average correlation (Spearman R) between splicing factor expression and isoform inclusion levels for each isoform cluster defined in E. Displayed are the top 35 splicing factors as ranked by highest absolute correlation values. For the box plots in D, G, H, box limits extend from the 25th to 75th percentile, while the middle line represents the median. Whiskers extend to the largest value no further than 1.5 times the inter-quartile range (IQR) from each box hinge. Points beyond the whiskers are outliers. Source data are provided as a Source Data file.