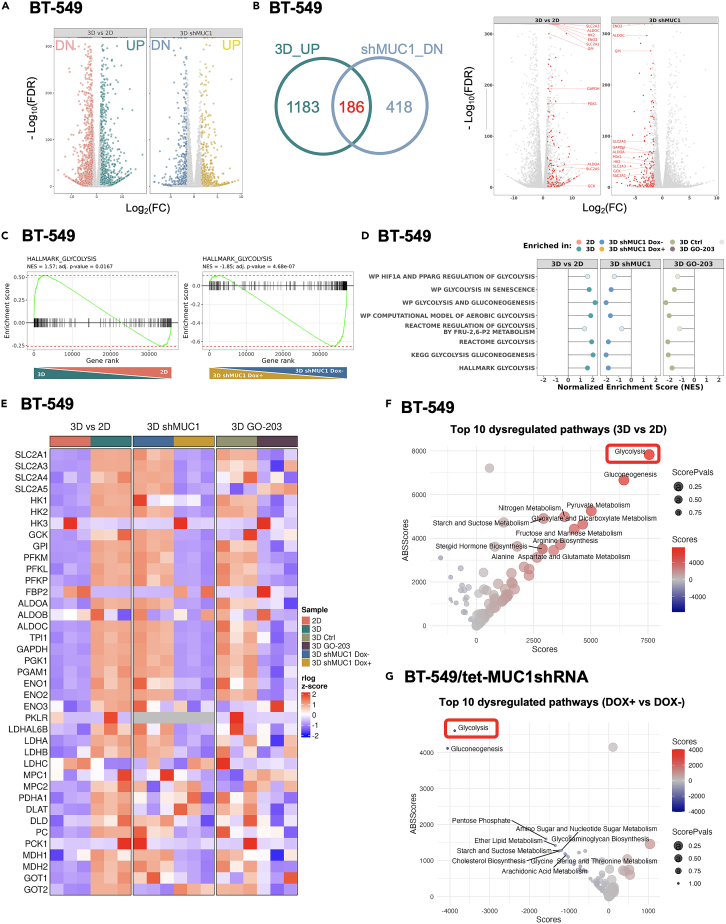
Figure 2.
MUC1-C is necessary for activation of glycolysis gene signatures in CSCs
(A) Volcano plots showing downregulated (left) and upregulated (right) genes in BT-549 cells grown in S6 3D vs. 2D culture (left; 3070 DEGs; <0.1 padj (Benjamini-Hochberg adjusted p value), log2fold change (FC) > 1.5), with 1,369 upregulated and 1,701 downregulated genes). Volcano plots of downregulated (left) and upregulated (right) genes in BT-549/tet-MUC1shRNA 3D cells treated with DOX vs. vehicle for 10 days (right; 1678 DEGs; <0.1 padj (Benjamini-Hochberg adjusted p value), log2fold change ((FC) > 1.5), with 1,074 upregulated and 604 downregulated genes).
(B) Overlap of DEGs upregulated in 3D BT-549 cells and downregulated in 3D BT-549/tet-MUC1 shRNA cells treated with DOX (left). The differential expression of these shared genes (red) is shown for the two conditions (right). Glycolytic genes are labeled within the shared set.
(C) GSEA of genes in BT-549 cells grown in 3D vs. 2D culture (left) and BT-549/tet-MUC1shRNA 3D cells treated with DOX vs. vehicle control (right) using the HALLMARK_GLYCOLYSIS gene signature. NES: Normalized Enrichment Score.
(D) GSEA lollipop plots of multiple glycolysis-related gene sets for (i) BT-549 cells grown in 3D vs. 2D culture and (ii) BT-549/tet-MUC1shRNA 3D cells treated with DOX vs. vehicle control.
(E) Heatmaps of glycolytic genes in (i) BT-549 cells grown in 3D vs. 2D culture, (ii) BT-549/tet-MUC1shRNA 3D cells treated with DOX vs. vehicle, and (iii) BT-549 3D cells treated with GO-203 vs. vehicle.
(F and G) MetaPhOR analysis identifying transcriptional dysregulation of metabolic pathways in 3D vs. 2D BT-549 cells (F) and 3D BT-549/tet-MUC1shRNA cells treated with DOX vs. vehicle (G).