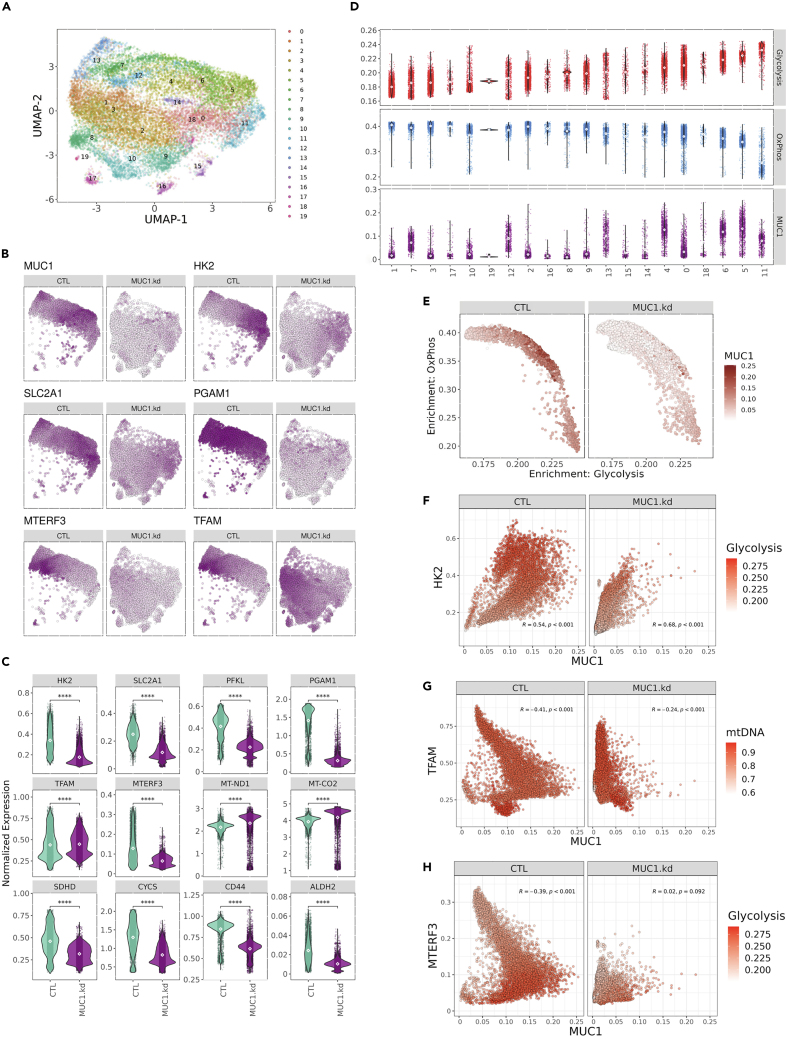
Figure 7.
Single-cell RNA-seq identifies metabolic heterogeneity in mammosphere CSCs that associates with MUC1 expression
(A) UMAP visualization of cells and clusters identified.
(B) Expression feature plots of select genes associated with glycolysis (HK2, SLC2A1, and PGAM1) and transcriptional regulators of mitochondrial DNA (TFAM and MTERF3) visualized by UMAP representation reveal metabolic heterogeneity within passaged sphere cells.
(C) Expression of glycolytic genes (HK2, SLC2A1, PFKL, and PGAM1), mitochondrial transcriptional regulators (TFAM and MTERF3), mitochondrial encoded components of ETC (MT-ND1 and MT-CO2), nuclear encoded components of ETC (SDHD and CYCS), and stemness genes (CD44 and ALDH2) associate with MUC1 status in passaged sphere cells.
(D) Metabolic signature scores and MUC1 expression levels across clusters.
(E) Comparison of single-cell HALLMARK_GLYCOLYSIS and HALLMARK_OXIDATIVE_PHOSPHORYLATION signature scores within individual cells. Color represents MUC1 expression within individual cells.
(F) Correlation of HK2 and MUC1 expression across CTL or MUC1 knockdown passaged sphere cells with color intensity representing HALLMARK_GLYCOLYSIS signature scores.
(G) Comparison of TFAM and MUC1 expression in CTL or MUC1 knockdown passaged sphere cells. Color represents overall signature of ETC genes encoded by the mitochondrial genome.
(H) Correlation of MTERF3 expression and MUC1 expression with color intensity indicating HALLMARK_GLYCOLYSIS enrichment score.