Fig. 1.
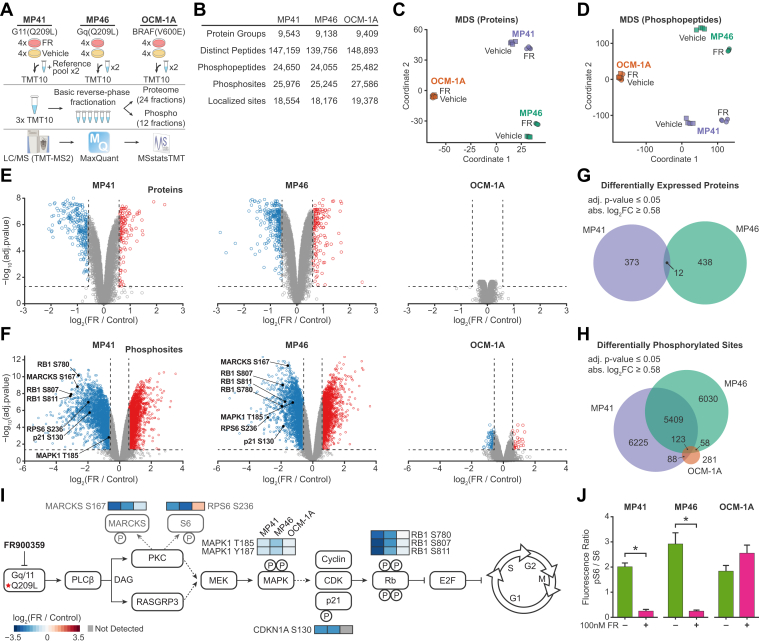
Sample comparisons show FR responses in Gq/11-driven UM cells.A, two PDX-derived Gq/11-driven UM cell lines, MP41(GNA11-Q209L) and MP46(GNAQ-Q209L), and an FR-insensitive UM cell line (OCM-1A) driven by BRAF(V600E) were treated with FR (four samples each) or vehicle (four samples each) for 24 h. Proteins were isolated, digested, and TMT-labeled prior to reverse-phase fractionation. Samples were analyzed by LC/MS1 and TMT-MS2 using the Q-Exactive mass spectrometer. The raw MS data files were searched with MaxQuant, and protein and phosphosite differential expression analyses were performed with the linear mixed-effects model from MSstatsTMT and MSstatsTMTPTM, using a moderated t-statistic, Benjamini-Hochberg multiple test correction, and normalization to protein abundance for phosphosites. B, protein and phosphosite yields are given for each cell line. Protein and phosphoprotein log2-ratios were aligned across samples so that the maximum likelihood of log-ratios was centered at zero for each sample and then normalized across all samples for metric multidimensional scaling (MDS) of proteins (C) or phosphopeptides (D) to assess differences among indicated groups of samples. Volcano plots were generated with the resulting adjusted p-values and estimated fold changes. E, volcano plots of protein changes in response to FR treatment were generated separately for each cell line. Blue indicates significant decreases (p < 0.05; log2 fold-change < −0.58; i.e. fold-change <50%) and red indicates significant increases (p < 0.05; log2 fold-change >0.58; i.e. fold-change >50%) in response to FR. F, volcano plots of changes in relative abundance of phosphosites in response to FR were generated for each cell line. Specific phosphosites relevant to Gq/11 signaling in MP41 and MP46 cell lines (I) are indicated. G, proteins were filtered by adjusted p-value <0.05 and absolute log2(fold-change) > 0.58 and compared among cell lines. Overlaps indicate significant changes among the indicated lines in the same direction in response to FR. H, phosphosites were filtered as indicated and compared among cell lines. Overlaps indicate significant changes in the same direction among the indicated lines. I, schematic diagram indicating the flow of signaling downstream of constitutively active Gq/11. Dotted lines indicate indirect interactions. Changes in phosphorylation of sites indicated in (F) are color-coded by Log2(fold-change) in response to FR. Constitutively active Gq/11 (Q209L) drives phospholipase Cβ (PLCβ) to produce diacylglycerol (DAG), which, in UM cells, activates both PKC and a Ras guanine nucleotide exchange factor (RASGRP3) that activate MAPK/Erk Kinase (MEK) to phosphorylate MAPK. Active MAPK drives cell proliferation through cyclin-dependent kinases (CDKs), which phosphorylate and inactivate the Rb family of cell cycle checkpoint inhibitors. J, immunoblots (supplemental Fig. S8) were performed to measure independently the phosphorylation of S6 (RPS6) on serine-246 (indicated in F and I) in response to FR in each cell line and showed significantly reduced (∗ p < 0.01) phosphorylation in MP41 and MP46 cells. UM, uveal melanoma.