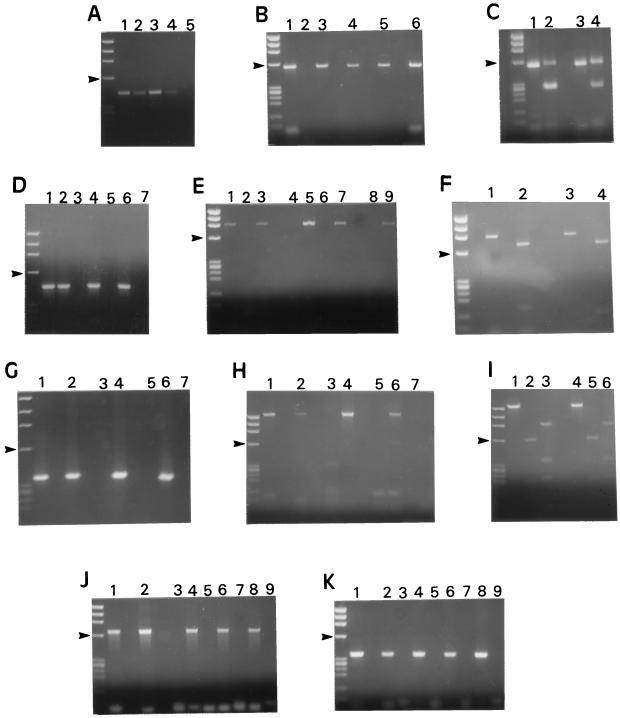
FIG. 1.
Ethidium bromide-stained agarose gels of PCR products amplified from C. parvum, C. muris, C. baileyi, and C. meleagridis DNA. Size markers are HaeIII-digested ΦX174; arrowheads in all panels point to the 603-bp fragment. (A) Amplification of a 435-bp product of the 18S rRNA gene (method of Johnson [13]). Lanes: 1, C. parvum; 2, C. meleagridis; 3, C. muris; 4, C. baileyi isolate B1; 5, negative control. (B) Amplification of a 556-bp product of the 18S rRNA gene (Awad-El-Kariem [1]). Lanes: 1, C. parvum; 2, negative control; 3, C. meleagridis; 4, C. baileyi isolate B1; 5, C. baileyi isolate O.96.2; 6, C. muris. (C) Electrophoresis of PCR products obtained in the experiment shown in panel B, prior to and after MaeI treatment. As expected (1), incomplete digestion occurred. Lanes: 1, C. parvum; 2, same sample as in lane 1 but after restriction with MaeI; 3, C. meleagridis; 4, same sample as in lane 3 but after restriction with MaeI. (D) Amplification of a 452-bp product of an undefined gene (Laxer [15]). Lanes: 1, C. parvum; 2, C. meleagridis; 3 and 4, C. muris; 5 and 6, C. baileyi isolate B1; 7, negative control. In lanes 4 and 6, C. parvum DNA was added to the reaction mixtures prior to amplification. (E) Amplification of a 898-bp fragment from the CpR1 gene (Wagner-Wiening [28]). Lanes: 1, C. parvum; 2, negative control; 3, C. meleagridis; 4 and 5, C. baileyi isolate B1; 6 and 7, C. baileyi isolate O.96.2; 8 and 9, C. muris. For lanes 5, 7, and 9, C. parvum DNA was added to the reaction mixtures prior to amplification. (F) Electrophoresis of PCR products obtained in the experiment represented in panel E prior to and after treatment with BamHI. Lanes: 1, C. parvum; 2, same sample as in lane 1 but after restriction with BamHI; 3, C. meleagridis; 4, same sample as in lane 3 but after restriction with BamHI. (G) Amplification of a 358-bp fragment of the CpR1 gene (Laberge [14]). Lanes: 1, C. parvum; 2, C. meleagridis; 3 and 4, C. muris; 5 and 6, C. baileyi isolate B1; 7, negative control. For lanes 4 and 6, C. parvum DNA was added to the reaction mixtures prior to amplification. (H) Amplification of a 1,500-bp fragment from an undefined DNA region (Bonnin [3]). Lanes: 1, C. parvum; 2, C. meleagridis; 3 and 4, C. muris; 5 and 6, C. baileyi isolate B1; 7, negative control. For lanes 4 and 6, C. parvum DNA was added to the reaction mixtures prior to amplification. (I) Electrophoresis of PCR products obtained from the 1,500-bp fragment shown in panel H prior to and after treatment with HinfI and RsaI. Lanes: 1, C. parvum; 2 and 3, same product as in lane 1 but after digestion with HinfI and RsaI, respectively; 4, C. meleagridis; 5 and 6, same product as in lane 4 after digestion with HinfI and RsaI, respectively. (J) Amplification of a 680-bp fragment from an undefined gene (Morgan [18]). Lanes: 1, C. parvum; 2, C. meleagridis; 3 and 4, C. muris; 5 and 6, C. baileyi isolate B1; 7 and 8, C. baileyi isolate O.96.2; 9, negative control. For lanes 4, 6, and 8, C. parvum DNA was added to the reaction mixtures prior to amplification. (K) Amplification of a 361-bp fragment of the Hsp70 gene (Rochelle [24]). Lanes: 1, C. parvum; 2, C. meleagridis; 3 and 4, C. muris; 5 and 6, C. baileyi isolate B1; 7 and 8, C. baileyi isolate O.96.2; 9, negative control. For lanes 4, 6, and 8, C. parvum DNA was added to the reaction mixtures prior to amplification.