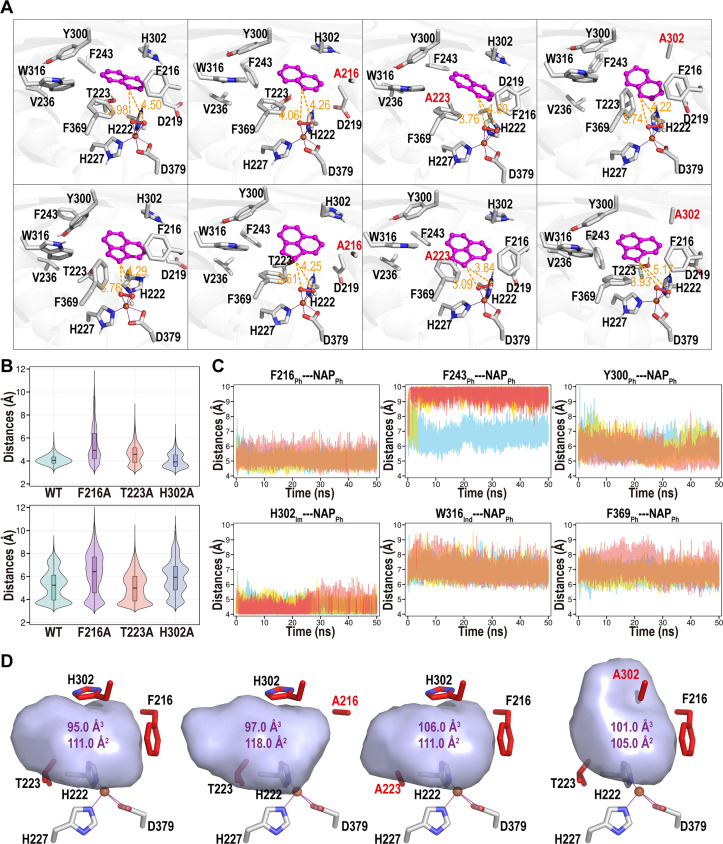
Fig 8.
Structural analysis of NarA2B2 complex with representative substrate. (A) Clustering structures of NarA2B2 and its mutations with NAP (top row) and ACE (bottom row). Active site residues are displayed in stick format, along with the substrate and OOH moiety, shown in stick and ball formats, respectively. Substrates (NAP and ACE) are highlighted in magenta. Attacking distances between the substrate and the OOH moiety are indicated in orange, with distances in Å. (B) Violin density plot illustrating the attacking distances during MD simulations between NarA2B2 and its mutations with either NAP (top) or ACE (bottom). The attacking distances are determined by the minimum distance between the carbon atom of the substrate and the oxygen atom bound to iron. (C) Potential π interaction distances between residues and NAP within the WT-NAP system complex. Distances are measured from the benzene ring’s center of mass within residues and NAP. The terms “Ph,” “Im,” and “Ind” denote phenyl, imidazole, and Indole rings, respectively. Data includes three parallel MD simulations, represented by red, yellow, and blue lines. (D) Active site pocket structures of NarA2B2 and its mutations, with calculated volumes (in Å3) and areas (in Å2).