ABSTRACT
The evolution of a unique craniofacial complex in vertebrates made possible new ways of breathing, eating, communicating and sensing the environment. The head and face develop through interactions of all three germ layers, the endoderm, ectoderm and mesoderm, as well as the so-called fourth germ layer, the cranial neural crest. Over a century of experimental embryology and genetics have revealed an incredible diversity of cell types derived from each germ layer, signaling pathways and genes that coordinate craniofacial development, and how changes to these underlie human disease and vertebrate evolution. Yet for many diseases and congenital anomalies, we have an incomplete picture of the causative genomic changes, in particular how alterations to the non-coding genome might affect craniofacial gene expression. Emerging genomics and single-cell technologies provide an opportunity to obtain a more holistic view of the genes and gene regulatory elements orchestrating craniofacial development across vertebrates. These single-cell studies generate novel hypotheses that can be experimentally validated in vivo. In this Review, we highlight recent advances in single-cell studies of diverse craniofacial structures, as well as potential pitfalls and the need for extensive in vivo validation. We discuss how these studies inform the developmental sources and regulation of head structures, bringing new insights into the etiology of structural birth anomalies that affect the vertebrate head.
Keywords: Craniofacial, Cranial neural crest, Single-cell genomics, Vertebrate head development, Congenital anomalies
Summary: This Review summarizes recent insights into the development, evolution and diseases of the vertebrate head based on single-cell profiling of gene expression and regulatory elements across diverse craniofacial tissues.
Introduction
Craniofacial development involves an intricate series of morphogenetic processes in the embryonic head. Shortly after gastrulation, cranial neural crest cells (CNCCs) emerge from the neural plate border through an epithelial-mesenchymal transition and migrate throughout the head. At around the same time, the pharyngeal endodermal epithelium forms a series of evaginations (pouches), which contact ectodermal epithelial invaginations (clefts). Within each pharyngeal arch segment, the pouch and cleft epithelia envelop CNCC mesenchyme and a core of mesoderm-derived mesenchyme (Fig. 1A) (Chai and Maxson, 2006).
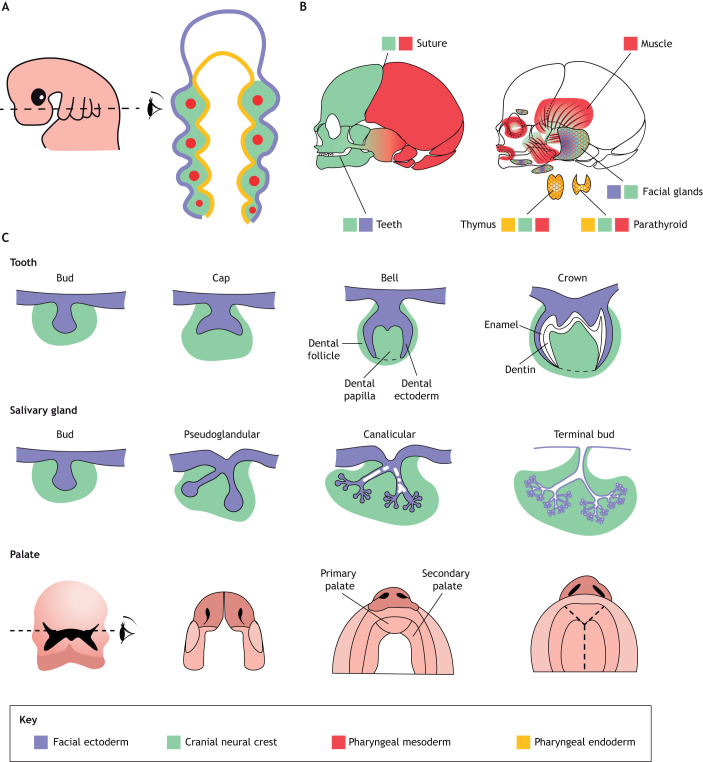
Fig. 1.
Overview of human craniofacial development and organ systems. (A) Pharyngeal arch stage (human 4-5 weeks) with cross-sectional diagrams illustrating the major germ layer contributions. Facial ectoderm, purple; cranial neural crest, green; pharyngeal mesoderm, red; pharyngeal endoderm, orange. (B) Contribution of germ layers to the bones and teeth of the face and skull (left), muscles and glands (right). (C) Developmental stages of a typical tooth, facial gland and palate. Germ layer contributions are shown for the tooth and gland.
The three primary germ layers (ectoderm, endoderm and mesoderm), along with the cranial neural crest (often referred to as the fourth germ layer), contribute to distinct cell types and communicate with each other to pattern craniofacial structures (Fig. 1B). CNCCs contribute to musculoskeletal structures (bone, cartilage, teeth, tendons and ligaments), as well as smooth muscle and fibroblasts (Fabian et al., 2022). Meanwhile, the mesoderm generates the facial muscles and some parts of the head skeleton, particularly the posterior skull, the endoderm contributes to endocrine organs such as the thymus and parathyroid, and the ectoderm contributes to the salivary and lacrimal glands (Lescroart et al., 2015; Rothova et al., 2012). Over a century of dye labeling, tissue grafting and genetic recombination experiments have revealed extensive catalogs of cell types derived from each germ layer (Fabian and Crump, 2022).
Identification of the genes and signaling pathways required for craniofacial development has come from multiple approaches. Forward genetic screens in model organisms such as zebrafish and mice have identified many genes required for craniofacial development (Amsterdam et al., 2004; Piotrowski et al., 1996; Schilling et al., 1996), while genome-wide association studies (GWAS) have identified candidate genes in humans with congenital anomalies (e.g. cleft palate and craniosynostosis) (Justice et al., 2020; Leslie et al., 2017). Reverse genetic approaches test the requirements of candidate genes and pathogenicity of mutations through genetic engineering of mouse embryonic stem cells and diverse model organisms (Van Otterloo et al., 2016). Conditional genetics, in which genes are deleted or modified in specific tissue types (e.g. through Cre-mediated recombination), reveal in which tissues genes function for craniofacial development (Murray, 2011).
Despite these advances, we have an incomplete picture of the genetic and mechanistic underpinnings of craniofacial development. Genetic studies often miss key genes involved in craniofacial development due to genetic redundancy, pleiotropy and species-specific requirements. Changes in gene regulation play major roles in congenital anomalies and evolution, yet our ability to understand the non-coding regulatory genome is only just emerging. Recent advances in high-throughput sequencing, including at the single-cell level, have enabled a more holistic view of the repertoire of expressed genes and active gene regulatory elements in diverse cell types of the developing head. In this Review, we discuss the opportunities and challenges in leveraging these massive datasets to make new insights into craniofacial development, disease and evolution.
Single-cell studies of craniofacial germ layers
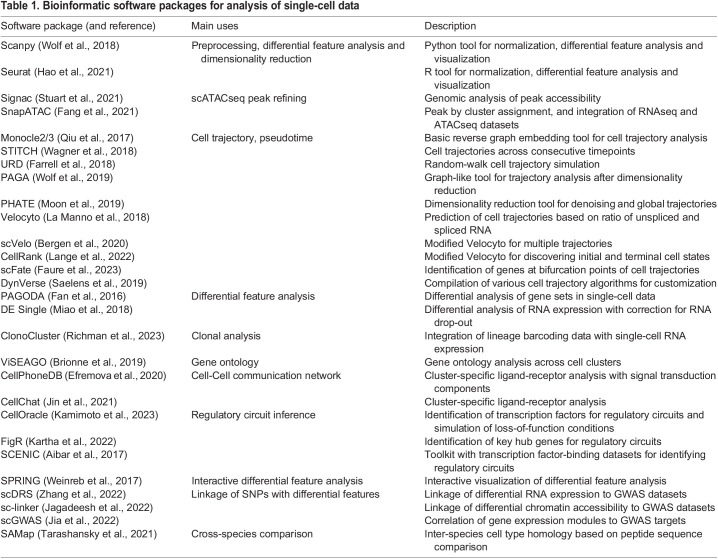
Cells can be classified into types based on functions, morphologies, locations, protein compositions and other properties. High-throughput DNA sequencing, combined with the ability to uniquely barcode single cells, has led to new opportunities to define all cell types within a tissue, organ or entire organism. Current approaches include single-cell or single-nuclei sequencing of reverse transcribed RNA molecules (scRNAseq) or regions of the genome accessible to transposase integration (snATACseq) (Buenrostro et al., 2015) (Fig. 2B). Accessible regions include the chromatin-depleted regions of enhancers, promoters, insulators and other gene regulatory domains. scRNAseq and snATACseq datasets can be integrated using software [e.g. SnapATAC; (Fang et al., 2021)] that uses accessibility of gene loci in snATACseq data to generate pseudotranscriptomic data, or by performing multiomic experiments on the same barcoded cells (Cao et al., 2018). Lineage information can then be deduced by connecting cells with overlapping gene expression (e.g. Monocle 3; Cao et al., 2019) or by comparing nascent RNA versus spliced RNA to infer the trajectory of gene expression (e.g. RNA Velocity; La Manno et al., 2018). Gene regulatory networks can be predicted by constructing regulons of correlated gene expression (e.g. SCENIC; Aibar et al., 2017) or by linking expression of transcription factors with their binding site enrichment in accessible chromatin domains (e.g. CellOracle; Kamimoto et al., 2023). Receptor and ligand expression can also be used to identify potential cell-cell signaling pathways [e.g. CellChat (Jin et al., 2021) and CellPhoneDB (Efremova et al., 2020)] (Fig. 2C). See Table 1 for a summary of single-cell bioinformatics software packages.
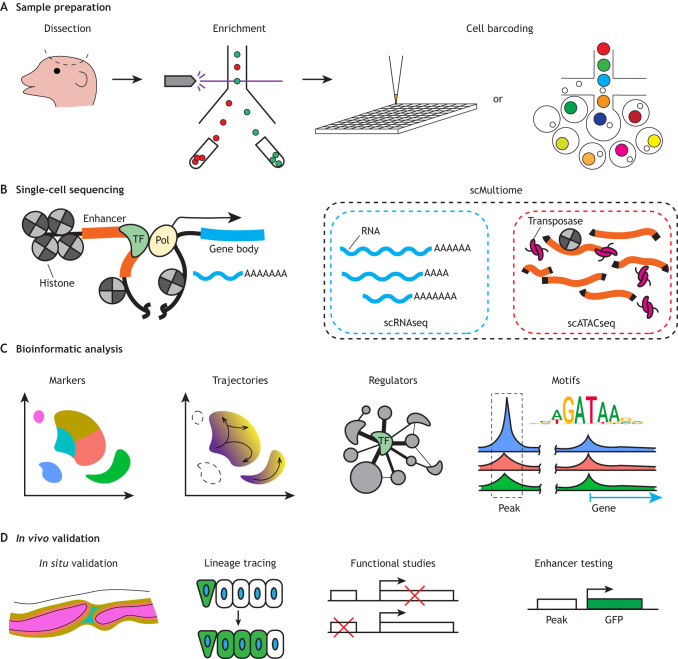
Fig. 2.
Flow chart of single-cell studies from sample preparation to validation. (A) Craniofacial tissues can be dissected, and specific cell types can be enriched by fluorescence-activated cell sorting (FACS) based on transgenes or cell surface markers. Cell barcoding can be performed by sequential pipetting into multi-well plates with unique barcodes or by emulsion with barcode-containing lipid droplets. (B) Diagram of a transcriptional locus showing sources of RNA for scRNAseq and accessible chromatin for scATACseq. (C) Bioinformatics packages can be used to generate cell clusters and marker genes, to infer potential cell lineage trajectories, and to identify potential master regulatory genes and potential enhancers. (D) In vivo validation includes mRNA in situ hybridization, lineage tracing, loss- and gain-of-function studies, and transgenic testing of enhancers.
Table 1.
Bioinformatic software packages for analysis of single-cell data
While these approaches are powerful, they are also prone to misinterpretation due to batch effects (Büttner et al., 2019; De Rop et al., 2023), over -or under-clustering of data (Xia and Yanai, 2019), limitations of lower dimensional space visualization tools such as tSNE and UMAP (Chari and Pachter, 2023), and arbitrary parameter selection in trajectory analyses (Gorin et al., 2022; Weinreb et al., 2018). In all cases, these approaches simply make predictions about the underlying biology that must be carefully validated in vivo (Fig. 2D).
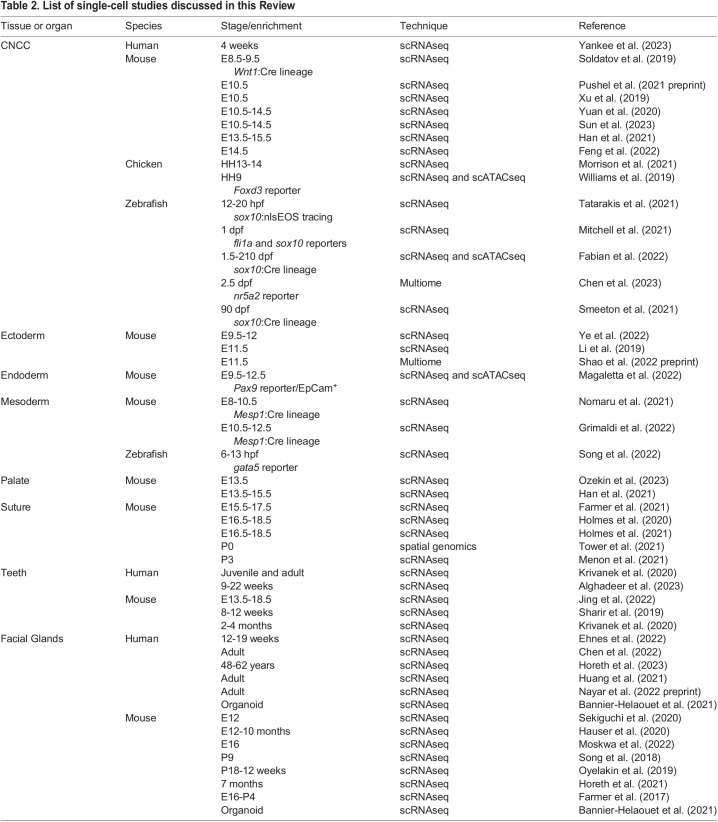
Two major approaches have been taken to isolate craniofacial tissues for single-cell profiling (Fig. 2). The first involves dissection of particular structures, such as the maxillary and mandibular prominences, or the teeth, sutures, glands and palate during later development (Fig. 2A). This dissection approach has been applied largely to mouse and, to a limited extent, human embryos (Yankee et al., 2023). The second approach involves cell sorting to enrich specific cell populations or lineages. Cell types can be purified based on active expression of reporter transgenes or surface proteins. Alternatively, cells of a particular lineage can be labeled by permanent genetic recombination earlier in development. One of the most common approaches in mouse is the Cre-Lox system (Murray, 2011), where tissue-specific Cre recombinase excises a LoxP-flanked stop cassette to allow expression of a reporter gene from a ubiquitous promoter in all derivatives of the Cre-expressing cells. Combined approaches can also be taken, such as isolating CNCC-lineage cells labeled by the Cre-Lox system specifically from the dissected jaw. Marker expression can also be used to extract craniofacial cell types from larger single-cell datasets, such as those from the Human Cell Atlas Project (Caetano et al., 2022). Despite the high prevalence of craniofacial anomalies, craniofacial cell types are not a major focus of current cell atlas projects and can be rare in whole-animal datasets, necessitating more-focused single-cell studies (Table 2).
Table 2.
List of single-cell studies discussed in this Review
Cranial neural crest cells
Since the discovery that CNCCs generate cartilage and teeth in the skull (Platt, 1893), researchers have used various methods to catalog the diversity of CNCC-derived cell types (see reviews by Fabian and Crump, 2022; Tang and Bronner, 2020). Methods include dye labeling in amphibia (Collazo et al., 1993) and avian chimeras (Le Lievre and Le Douarin, 1975), as well as genetic recombination-based tracing, primarily Wnt1-Cre in mouse (Jiang et al., 2002) and Sox10-Cre in zebrafish (Kague et al., 2012). A unique feature of CNCCs, as opposed to trunk neural crest, is their ability to form not only neuroglial and pigment cell types, but also the ectomesenchyme that contributes to bone, cartilage, teeth and other connective tissues (Le Lievre and Douarin, 1975). How CNCCs generate such diverse cell types with spatiotemporal precision remains a primary focus of the field.
In chick, scRNAseq, snATACseq and bulk epigenomic profiling data suggest that CNCCs are biased towards ectomesenchyme versus neuroglial lineages at pre-migratory stages (Williams et al., 2019). However, a separate study concluded that heterogeneity in migratory chick CNCCs corresponds more to leader versus follower migratory behavior, rather than to lineage potential (Morrison et al., 2017). In mouse, scRNAseq of pre-migratory and migratory CNCCs at embryonic day (E) 8.5 and E9.5 identified migratory CNCCs with shared mesenchymal and neuroglial programs, which resolved into distinct ectomesenchymal and neuroglial programs through cross-repression (Soldatov et al., 2019). In zebrafish, photoconversion of a sox10:nlsEOS line was used to label and sort mandibular CNCCs for scRNAseq analysis (Tatarakis et al., 2021). As in mouse, this study revealed that the earliest lineage decisions of zebrafish CNCCs into ectomesenchyme, pigment and neuroglial fates occurs at late migration stages. These latter studies agree with genetic evidence that ectomesenchyme identity is established during CNCC migration, coincident with downregulation of early CNCC specifiers sox10 and foxd3, and upregulation of ectomesenchyme markers twist1a, dlx2a and fli1a (Bildsoe et al., 2009; Blentic et al., 2008; Das and Crump, 2012).
Upon migration into the pharyngeal arches, CNCC ectomesenchyme acquires positional information. Anterior-posterior identity is retained from CNCC origins near the nascent midbrain and hindbrain (Hunt et al., 1991), with the hyoid and more-posterior branchial arches, but not the mandibular arch and frontonasal prominence, expressing anterior Hox genes (Miller et al., 2004). Signaling from the arch epithelia regionalizes CNCCs along the dorsoventral (i.e. proximodistal), mediolateral and oral-aboral axes (reviewed by Minoux and Rijli, 2010), as exemplified by epithelial-derived endothelin 1 induction of nested Dlx gene expression in CNCCs along the dorsoventral axis (Beverdam et al., 2002; Depew et al., 2002; Talbot et al., 2010). In zebrafish, bulk transcriptomic analyses comparing fli1a:GFP+; sox10:dsRed+ CNCC ectomesenchyme with CNCCs from dlx5a:GFP+ or hand2:GFP+ dorsoventral subdomains identified cohorts of region-specific CNCC genes (Askary et al., 2017). Subsequently, scRNAseq analysis of fli1a:GFP+; sox10:dsRed+ arch CNCCs showed that, before differentiation, CNCCs are largely distinguished by regional identity along the anteroposterior axis (Mitchell et al., 2021). Additionally, integration of scRNAseq and snATACseq data of zebrafish post-migratory CNCCs, labeled by Sox10:Cre, revealed clustering to be driven by anteroposterior, dorsoventral and oral-aboral identity (Fabian et al., 2022). The strong effect of regional identity on clustering allowed researchers to create a virtual three-dimensional map of pharyngeal gene expression, which predicted with ∼90% accuracy the precise arch expression domains of CNCC genes. For example, this analysis identified a previously unreported role for the nuclear receptor Nr5a2 in promoting tendon, ligament and glandular fates in the mandibular arches of zebrafish and mouse, with multiome analysis of fish nr5a2 mutants identifying a number of directly regulated jaw-specific enhancers (Chen et al., 2023).
In mouse, scRNAseq of dissected facial domains allowed profiling of the maxillary and mandibular prominences, and hyoid arch (Pushel et al., 2021 preprint; Sun et al., 2023; Xu et al., 2019; Yuan et al., 2020). Although dissected tissue contains cell types of each germ layer, CNCCs often make the largest contribution and can be subclustered based on ectomesenchyme gene expression. As in zebrafish, scRNAseq analysis of the mouse mandibular process at E10.5 revealed CNCC clustering driven by proximodistal and oral-aboral patterning (Pushel et al., 2021 preprint; Xu et al., 2019; Yuan et al., 2020). Analysis of co-regulated genes suggested that Hedgehog and Bmp signaling promote oral and aboral development, respectively, which was validated through genetic manipulation of both pathways in mouse embryos (Xu et al., 2019). scRNAseq of the mandibular prominences from E10.5-E14.5 revealed progressive fate restriction of CNCCs into fibroblastic and progenitor fates, with progenitors further bifurcating along bone and tooth lineages (Yuan et al., 2020). During the same period in the maxillary prominences, CNCCs bifurcate into palatal and tooth lineages (Sun et al., 2023). In chick, scRNAseq of each of the first four branchial arches revealed both a core migratory signature and arch-specific gene expression (Morrison et al., 2021). In human embryos, scRNAseq of the facial prominences at Carnegie stage 20 (Yankee et al., 2023), along with chromatin accessibility and histone mark data from multiple stages (Wilderman et al., 2018), showed strong correlation of cell types between human and mouse, as well as potential human-specific gene expression. Genes expressed in the developing human face were also enriched for known disease-causing mutations. In addition to revealing common signatures of arch regionalization across vertebrates, these studies reveal candidate genes and regulatory sequences that may be sites of damaging mutations in craniofacial anomalies.
Although comprehensive single-cell analyses of CNCC derivatives in adult mice has not been reported, scRNAseq and snATACseq have been used to profile zebrafish Sox10:Cre-labeled CNCC derivatives from pharyngeal arch to adult stages (Fabian et al., 2022). In addition to musculoskeletal tissue types, this study revealed CNCC-derived gill cell types unique to fish, including a specialized endothelial-like pillar cell for gas exchange (Mongera et al., 2013). Gills consist of several primary and secondary filaments that support respiration, with each filament arising clonally from a single CNCC (Stolper et al., 2019). Pseudotime analysis using Monocle3 revealed a putative fgf10b+ CNCC gill progenitor, which was confirmed as a progenitor for multiple gill cell types by photoconversion-based lineage tracing (Fabian et al., 2022). In contrast, pseudotime analyses of skeletal and connective tissue lineages generated poorly resolved trajectories, consistent with other studies (Bandyopadhyay et al., 2008; Colnot et al., 2004; Matsushita et al., 2020). One possibility is that skeletal and connective tissue progenitors exist in a continuum of states, with plasticity in response to homeostatic and regenerative needs. Furthermore, in vivo validation will be needed to resolve mesenchymal lineage relationships, likely aided by emerging barcoding and in situ readout technologies such as MEMOIR (Frieda et al., 2017).
Facial mesoderm
In addition to generating muscles and vasculature, the facial mesoderm generates many of the same connective tissue subtypes as CNCCs, including osteoblasts, chondrocytes, tendons, ligaments and fibroblasts (Grimaldi and Tajbakhsh, 2021). CNCCs and mesoderm both contribute in a seamless fashion to the skull bones (Koyabu et al., 2012; Teng et al., 2019), stapes cartilage of mouse (Thompson et al., 2012), otic cartilage of fish (Kague et al., 2012) and gill arch cartilage of rays (Sleight and Gillis, 2020). Despite these similar contributions, the differentiation of mesoderm into head connective tissues remains less understood than for CNCCs. To address this, scRNAseq of mouse Mesp1-Cre-labeled facial mesoderm and Myf5-Cre-labeled mesoderm was recently performed at E14.5 (Grimaldi et al., 2022). By analyzing cohorts of co-expressed genes using SCENIC (Aibar et al., 2017), potential drivers of the connective tissue lineage were identified, including genes shared with CNCC ectomesenchyme, such as Twist1 (Bildsoe et al., 2009; Das and Crump, 2012), Prrx1 (Barske et al., 2016; ten Berge et al., 1998), Pdgfra (Weston et al., 2004), Foxp2 (Chen et al., 2023), Fli1 (Das and Crump, 2012) and Six2 (Liu et al., 2019). Inhibition of the myogenic program by Myf5 loss resulted in increased contributions of facial mesoderm to connective tissue fates, and pseudotime analysis implicated Foxp2 as a potential regulator of connective tissue commitment (Grimaldi et al., 2022). In addition, scRNAseq of Mesp1-Cre- and Tbx1-Cre-labeled mesoderm, combined with bulk ATACseq and Tbx1 ChiP-Seq, revealed a central role for Tbx1 in opening and activating enhancers for cardiac and branchial muscle fates in a common progenitor (Nomaru et al., 2021). In zebrafish, scRNAseq analysis of gata5:GFP+ mesoderm indicated bifurcations between cardiac and facial connective tissue fates, with loss of Gata5/6 function skewing lineages toward facial connective tissue fates (Song et al., 2022). These findings highlight similarities between commitment of mesoderm into myogenic versus connective tissue lineages and of CNCCs into neuroglial versus ectomesenchyme connective tissue lineages, potentially reflecting convergence of mesoderm and CNCC lineage cells onto a mesenchyme subtype with similar connective tissue potential.
Pharyngeal endoderm
The pharyngeal endoderm forms a series of pouches that help pattern the craniofacial skeleton and contribute to several endocrine organs (Choe et al., 2013; Graham et al., 2005). In mouse, sorting of Pax9-Venus/EpCam+ cells from dissected pharynxes allowed the authors to catalog gene expression (by scRNAseq) and chromatin accessibility (by snATACseq) in pharyngeal endoderm cells from E9.5-E12.5 (Magaletta et al., 2022). Cell clustering on the integrated datasets revealed molecular differences between the first and second endodermal pouch and captured the emergence of the thyroid, parathyroid, ultimobranchial body and thymus. In addition to identifying previously unreported gene expression in these developing glands, single-cell analysis revealed the earliest markers of distinct medullary and cortical regions of the thymic epithelium. In silico perturbations of gene function by CellOracle were used to predict a key role for Foxn1 in the medullary thymic lineage, which has since been functionally validated in mice through knockout studies (Kadouri et al., 2022; Li et al., 2023).
In zebrafish, scRNAseq analysis of the adult pituitary, in which endodermal lineage cells were marked by sox17:CreER-mediated recombination, revealed an unexpected contribution of endodermal cells to the pituitary (Fabian et al., 2020). The endocrine component of the pituitary has classically been thought to derive from an ectodermal placode, the adenohypophysis, yet scRNAseq analysis revealed that endodermal cells contributed to all endocrine cell types of the pituitary, as validated by time-lapse imaging, analysis of multiple endoderm-labeling transgenes and mRNA in situ hybridization. As nonvertebrate chordates develop an endoderm-derived pituitary-like structure (Schlosser, 2017), the presence of endodermal lineage cells in the zebrafish pituitary may help clarify the origin and evolution of the vertebrate pituitary gland.
Facial ectoderm
As with pharyngeal endoderm, the facial ectoderm has an important signaling role in craniofacial patterning and contributes to diverse organs, including teeth, glands and taste buds. Epithelial cells, mostly of ectoderm origin, have been recovered in single-cell sequencing studies of the dissected murine maxillary prominence (Sun et al., 2023), mandibular prominence (Xu et al., 2019; Yuan et al., 2020) and hyoid arch (Pushel et al., 2021 preprint). Even after sorting based on CNCC-specific transgenes in zebrafish, epithelial cells are recovered, likely owing to the imperfect nature of the sorting process (Fabian et al., 2022; Tatarakis et al., 2021). In most of these studies, ectoderm lineage cells represented a small fraction of sequenced cells and were not analyzed in depth.
Ectoderm lineage cells of the frontonasal and maxillary prominences meet and fuse at the lambdoid junction and nasolacrimal groove to ensure proper palate and nasal development (Millicovsky et al., 1982). After dissection of these transient embryonic structures from E11.5 mice, scRNAseq of ectoderm lineage cells (marked by an ectoderm-specific Crect-Cre transgene) revealed that ectodermal derivatives include distinct basal epithelial cells within and outside the fusion sites, as well as dental epithelium, periderm and other cell types (Li et al., 2019). In a separate study, epithelial cells from the E12 mouse mandible were dissociated from the mesenchyme, sorted based on epithelial labeling by K14-Cre-mediated recombination and subjected to scRNAseq (Ye et al., 2022). As with CNCC mesenchyme, clustering of ectodermal epithelial cells was dictated largely by anatomical positions, including dental epithelia, adjacent posterior/oral and anterior/aboral epithelia, tongue epithelia with taste buds and the superficial periderm layer. Similar results were found by performing multiomics on mandibular and maxillary epithelia from E10.5 and E12.5 mouse embryos (Shao et al., 2022 preprint). In concordance with scRNAseq of the E12 mandible (Ye et al., 2022), the posterior/oral epithelium was distinguished by expression of Sox2, and Shh and Bmp signaling components, the anterior/aboral epithelium was distinguished by expression of Tfap2a, Tfap2b and Wnt signaling components and, at their intersection, the dental epithelium was distinguished by Pitx2 expression. Distinct transcription factor and signaling pathway expression in these domains was independently validated by bulk RNA sequencing after laser microdissection of domains from the E11.5 mandible (Shao et al., 2022 preprint), with both studies using in situ validation at multiple embryonic stages to show progressive establishment of distinct dental and non-dental epithelial domains. Interestingly, expression of some signaling molecules, such as Wnt10b, were detected in bulk RNAseq and scRNAseq, but not in multiome sequencing, highlighting recently reported tradeoffs in recovering lower abundance transcripts when combining sequencing modalities in single experiments (Booeshaghi et al., 2023 preprint). As discussed below, these studies provide insights into the earliest patterning events of the facial ectoderm that prefigure development of the palate, teeth, glands and other epithelial structures.
Single-cell studies of craniofacial organs
Palate
The primary palate forms from the anterior-most nasal processes, with the secondary palate zippering closed to separate the nasal and oral cavities. Development of the secondary palate involves medial growth of the maxillary processes, which elevate above the tongue to fuse with the lateral and medial nasal processes (Fig. 1C) (Burdi and Faist, 1967). Formation of the secondary palate requires involvement of multiple germ layers and signaling pathways (Compagnucci et al., 2021; Hammond and Dixon, 2022). scRNAseq of the dissected secondary palate in E13.5 mice revealed regionalization of the mesenchyme into Msx1+ proximal and Shox2+ oral domains (Ozekin et al., 2023), consistent with earlier expression studies (Han et al., 2009; Yu et al., 2005).
Although the hard palate consists of CNCC-derived bone, the soft palate consists of muscularized tissue that opens and closes the nasopharyngeal and oral cavities. scRNAseq of soft palate development in E13.5, E14.5 and E15.5 mice revealed a population of CNCC-derived perimysial cells that, through in situ validation, were confirmed to surround the developing palatal muscles (Han et al., 2021). A role in perimysial cell specification was identified for Runx2 (Han et al., 2021), which is well characterized in osteoblast formation (Ducy et al., 1997). Using CellChat, a potential role for transforming growth factor β (TGFβ) signaling in perimysial cell specification was also indicated (Feng et al., 2022), which was confirmed by Osr2-Cre-mediated deletion of Tgfbr1 or its target gene Fgf18 (Feng et al., 2022). These studies show the power of single-cell studies in identifying new and unexpected roles of genes and signaling pathways in CNCC development. Integrating single-cell and GWAS datasets can also point to cell populations and genetic pathways disrupted in cleft lip and palate, such as the Irf6-Tfap2a-Grhl3 gene circuit in the periderm (Siewert et al., 2023) – the outermost layer of the embryonic epithelium that serves to prevent inappropriate fusions of the palatal shelves during development (Richardson et al., 2014).
Teeth
Tooth development involves crosstalk between oral epithelia and CNCC mesenchyme, which induces differentiation of epithelial cells into enamel-producing ameloblasts and CNCCs into dentin-producing odontoblasts, pulp, periodontal ligament, cementum and alveolar bone (Fig. 1C) (Balic and Thesleff, 2015). Our understanding of the mechanisms behind the shaping and complex differentiation of teeth remains incomplete. scRNAseq of the E12 mouse mandibular epithelium, coupled with mRNA in situ hybridization, revealed previously unreported markers of the initiation knot, which is the signaling center for tooth initiation, including Gad1, Sp5 and Proser2 (Ye et al., 2022). One surprising finding was that Ntrk2, which helps transduce neurotrophic signaling in the nervous system, also plays a role in the proliferative growth and morphogenesis of incisors. Single-cell analysis also revealed the striking gene expression similarities of tooth germs and taste buds (Ye et al., 2022), consistent with experimental evidence in cichlid fish and mouse that taste buds can be partially converted to a tooth-like program by modulating Bmp signaling (Bloomquist et al., 2019).
Several groups have studied the mouse molar to understand how the diverse cell types derived from the CNCC mesenchyme arise. Jing et al. performed scRNAseq of the mouse molar from E13.5 to postnatal day (P) 7.5 (Jing et al., 2022). By E14.5, mesenchyme can be separated into the dental papilla, which generates the odontoblasts and pulp, and the dental follicle, which generates the periodontal ligament, cementum and alveolar bone. Furthermore, CellChat predicted roles for insulin-like growth factor (IGF) signaling within the dental follicle domain, which was validated by periodontal ligament defects upon Cre-mediated deletion of the Igf1 ligand or its receptor, Igf1r (Jing et al., 2022). These findings highlight the usefulness of the many publicly available Cre mouse lines to validate single-cell predictions, as well as to dissect cell-cell receptor-ligand interactions driving cell fate decisions.
In contrast to the molar, the rodent incisor is continuously growing, thus providing an opportunity to study stem cell regulation in both epithelial and mesenchymal compartments. One of the first global gene expression studies of teeth used microarray analysis in 94 dissected young adult mouse incisors to identify cohorts of co-varying gene modules (Seidel et al., 2017). In situ validation revealed multiple types of stem and progenitor populations in epithelial and mesenchymal compartments. In a subsequent study, scRNAseq of Epcam+ epithelial cells of the incisor revealed broad populations of proliferative progenitors that bifurcate to ameloblast and non-ameloblast lineages. Comparative scRNAseq of incisors during homeostasis and chemical injury-induced regeneration further revealed increased recruitment of Notch1-CreER-labeled stratum intermedium cells, which sit under the inner enamel epithelium, to regenerate ameloblasts (Sharir et al., 2019). This provides a good example of how lineages can be modified in response to injury to promote efficient repair.
Another approach to study the cellular composition of mouse incisors employed a combination of SMART-seq2, which provides high sequencing depth to relatively few cells, and 10x Chromium scRNAseq, which allows lower depth sequencing of many cells (Krivanek et al., 2020). In addition to confirming many of the independently reported progenitors, ameloblast intermediates and non-ameloblasts in the epithelium (Seidel et al., 2017; Sharir et al., 2019), a rare subtype of epithelial cell expressing mechanotransduction and calcium-dependent genes was identified. Whether this cell type functions to regulate incisor growth in response to mechanical forces remains to be corroborated. This same study also revealed different types of immune cells in the adult incisor, with implications for how the immune system may function to prevent caries (Krivanek et al., 2020). Comparative scRNAseq on the non-growing adult mouse molar revealed common signatures of shared tooth cell types, as well as unique cell types and gene expression that may underlie the ability of the incisor to continuously grow (Krivanek et al., 2020). Moreover, recent single-cell studies of developing human teeth, including non-growing incisors, also provide a foundation for comparisons with the continuously growing incisors of mice (Alghadeer et al., 2023).
Sutures
Sutures separate the calvarial bones and coordinate growth of the skull with protection of the underlying brain (Fig. 1B). In craniosynostosis, sutures fuse prematurely, leading to defective skull growth and neurocognitive defects (Twigg and Wilkie, 2015). Several studies have performed single-cell profiling of the sutures to understand the nature of the stem/progenitor and other cells that grow the bones and keep them separate. Holmes et al. profiled the embryonic mouse frontal suture, identifying osteoblasts, several clusters of suture-associated mesenchyme, overlying hypodermis, underlying dura, endothelial cells, pericytes and diverse immune cells (Holmes et al., 2020). In situ RNA hybridization revealed two populations of mid-suture mesenchyme: bone-associated osteoprogenitors and proliferative cells. Profiling of sutures in a mouse Twist1+/− model of Saethre-Chotzen syndrome, a genetic condition in which the coronal sutures are lost, and a Fgfr2S252W/+ model of Apert syndrome, in which multiple sutures are fused, revealed distinct changes in gene expression but no alterations to suture cell type composition (Holmes et al., 2020).
Two scRNAseq studies of the coronal suture identified largely similar cell types to those of the frontal suture (Farmer et al., 2021; Holmes et al., 2021). Both the ectocranial layers overlying the calvarial bones (Merrill et al., 2006; Ting et al., 2009) and the meninges situated between the calvaria and brain (Yu et al., 2021) are known to regulate suture formation and homeostasis. Farmer et al. revealed a diversity of ectocranial layers, including a ligament-like cell population bridging the suture that may have roles in mechanotransduction and/or stabilization of the suture (Farmer et al., 2021). Meanwhile, the dura contains a cartilage-like population that may have roles in suture closure through endochondral ossification. scRNAseq of suture chondrocytes in mice lacking Gnas, the mutation of which in humans is linked to fusion of multiple sutures, revealed their transformation into osteoblasts that closed the suture (Xu et al., 2022).
Comparative single-cell studies have the potential to reveal molecular differences between sutures that make them differentially sensitive to genetic and environmental perturbations. For example, one study identified unique expression of the Hedgehog signaling regulator Hhip in the coronal versus the frontal mid-suture region (Holmes et al., 2021). Lineage tracing with Hhip-CreER showed that Hhip+ mid-suture mesenchyme is not a major source of new osteoblasts in the short term, although there is modest contribution to calvarial osteoblasts after several months (Holmes et al., 2021). This result is similar to lineage tracing observed in mid-suture mesenchyme by Axin2-CreER (Maruyama et al., 2016), and differs from the extensive contribution of cells traced from both the bone fronts and mid-suture mesenchyme by Gli1-CreER (Zhao et al., 2015) or Ctsk-Cre (Debnath et al., 2018). These different findings suggest that new bone addition occurs mainly at the bone fronts, with mid-suture mesenchyme representing a reserve pool of cells that may contribute primarily during repair. As a note of caution, a scRNAseq study of cells from the frontal, coronal and sagittal sutures of mouse recovered two unique populations in the sagittal suture (Menon et al., 2021). However, closer examination suggests that these represent chondrocytes that were included preferentially in sagittal suture dissections. Thus, differences in dissection technique between sutures and between research groups need to be considered when analyzing cell compositional differences between suture stages and types.
A limitation of single-cell profiling is the inability to directly correlate cell clusters to spatial domains within the embryo. To address this, new ‘spatial transcriptomic’ methods have been developed to preserve spatial information by applying barcoding beads across tissue sections on slides. After sequencing, indexing the spatial position of beads allows mapping of global gene expression back to precise positions on the sectioned tissue (Ståhl et al., 2016). Such technology was applied to the sagittal sutures of newborn mice and gradients of gene expression from the mid-suture region to the flanking calvarial bones were analyzed (Tower et al., 2021). When cranial nerves innervating the suture were ablated, spatial transcriptomics revealed redistribution of Bmp and TGFβ pathway genes, which correlated with increased Bmp and TGFβ signaling in the mid-suture region and partial fusion of the suture. This analysis highlights the power of spatial genomics in revealing how changes in gene expression and cell distribution correlates to developmental defects of craniofacial structures.
Glands
The facial glands lubricate and protect the oral cavity and ocular surface. The submandibular and sublingual salivary glands produce mucous-rich secretions, whereas the parotid salivary gland produces watery secretions rich in digestive enzymes and the lacrimal gland secretes tears (de la Cuadra-Blanco et al., 2003; Martinez-Madrigal and Micheau, 1989). Major epithelial cell types include secretory acinar cells; intercalated, striated and excretory ductal cells that modify and conduct the secretions to the oral cavity or eye; and myoepithelial cells that promote secretion through contraction (Lombaert et al., 2013). Development of the glands shares properties with that of teeth, in particular interactions of CNCC mesenchyme with the overlying ectodermal epithelium to promote budding, branching and differentiation (Fig. 1C) (Grobstein, 1953; Jaskoll et al., 2002; Johnston et al., 1979; Rothova et al., 2012).
Single-cell RNA profiling has been performed on several glandular structures to characterize their development (Table 2). For example, Sekiguchi et al. compared the murine submandibular and parotid glands at E12 and found greater inter-gland differences in the mesenchymal than epithelial populations (Sekiguchi et al., 2020). The submandibular gland mesenchyme expressed a unique set of genes from parotid gland mesenchyme, including Nr5a2, which is essential for initiation and formation of the submandibular but not the parotid gland (Chen et al., 2023). Reciprocally, parotid gland mesenchyme displayed selective expression of the transcription factor Pou3f3 and associated long non-coding RNAs Pantr1 and Pantr2, potentially reflecting the role of Pou3f3 in development of the proximal arches where the parotid gland forms (Barske et al., 2020; Jeong et al., 2008). These mesenchymal expression differences may help establish differences in mucous versus aqueous secretions of these glands, with increased numbers of myoepithelial cells recovered in submandibular gland epithelia, reflecting the need for greater contractile forces in secretion of more viscous mucous (Tucker, 2007).
scRNAseq datasets for the mouse submandibular gland have been generated for bud (E12 and E14), branching (E16), postnatal (P1 and P30) and adult (10 month) stages, with a focus on epithelial cells (Hauser et al., 2020). Trajectory analysis using the dimensionality reduction algorithm PAGA (Wolf et al., 2019) suggests a late embryonic Krt19+ ductal population as the common progenitor for postnatal acinar, intercalated duct and striated duct cells. By performing scRNAseq of the P8 mouse submandibular gland, Song et al. identified a distinct SMA+ progenitor for basal epithelial and myoepithelial cells, which was confirmed with Acta2(SMA)-CreER-based lineage tracing (Song et al., 2018). Two distinct types of putative acinar progenitors, labeled by Smgc and Bpifa2, have also been identified that may give rise to distinct mucous and serous acinar cells, respectively (Hauser et al., 2020). Sexually dimorphic expression was also noted in adult glands, with Smgc, for example, expressed in female but not male mucous acinar cells. Sexual dimorphism has also been noted in scRNAseq studies of other epithelial organs, such as the kidney (Ransick et al., 2019), highlighting the importance of separately profiling male and female organs, especially at mature stages.
Although model organisms such as the mouse are valuable for uncovering core developmental processes of organs, there may also be human-specific gene expression and other features that are important for understanding disease etiology. As in mouse, scRNAseq of submandibular glands from 12- to 19-week-old human fetuses revealed KRT19+ basal epithelial cells as potential progenitors to all differentiated epithelial cell types, except for excretory ducts and myoepithelial cells (Hauser et al., 2020). Comparative studies also revealed similar cell compositions between human and mouse parotids (Chen et al., 2022), and minor salivary glands (Huang et al., 2021). Although serous acinar cells were similar between human and mouse (Chen et al., 2022), human mucous acinar cells were highly divergent from those in mouse, and no human equivalents of mouse granular convoluted tubular cells were evident (Horeth et al., 2023). These cellular differences may reflect divergent physiological properties of human versus mouse glands that may impact modeling glandular diseases in rodents.
Single-cell analysis has also been used to benchmark the fidelity of salivary gland (Moskwa et al., 2022) and lacrimal gland (Bannier-Helaouet et al., 2021) organoids; for example, showing that current lacrimal gland organoids are largely ductal in nature. Single-cell sequencing of mouse models (Horeth et al., 2021) and human tissues (Nayar et al., 2022 preprint) from individuals with Sjögren's syndrome, an autoimmune disease affecting facial gland function, revealed dynamic changes in immune and stromal cell components of the glands, as well as upregulation of immune regulators in glandular epithelial cells that may contribute to the disease. In addition, single-cell profiling of human minor salivary glands and oral mucosa revealed cell type-specific expression of the viral entry factor ACE2, which may explain increased susceptibility of these glands to COVID-19 infection (Huang et al., 2021).
Conclusions and future perspectives
Single-cell studies of craniofacial development generate massive amounts of data, yet most strategies to validate findings in vivo are low throughput. For RNA expression, new spatial transcriptomic approaches, such as MERFISH (Xia et al., 2019) and seqFISH (Coskun and Cai, 2016), allow the simultaneous detection of thousands of different transcripts in the same cell. When combined with spatial barcoding techniques such as MEMOIR (Askary et al., 2020; Frieda et al., 2017), these techniques could provide holistic lineage and spatial information for cell types, although the need for sectioning limits the ability to generate three-dimensional maps. To complement snATACseq predictions of enhancers, single-cell profiling of chromatin marks, DNA methylation, transcription factor occupancy and chromosome conformation could predict active, poised or repressed enhancers, as well as other types of boundary and regulatory elements. In vivo adaptation of high-throughput reporter assays, such as MPRA-seq (Inoue and Ahituv, 2015) and STARR-seq (Arnold et al., 2013), could also aid in dissecting the enhancer logic of craniofacial development.
Single-cell datasets can also be integrated with GWAS to identify potential causative mutations for craniofacial anomalies (Gawel et al., 2019; Jagadeesh et al., 2022; Jia et al., 2022; Zhang et al., 2022), including cleft palate (Leslie et al., 2016) and craniosynostosis (Justice et al., 2020). Given developmental gene conversation across vertebrates, coding mutations can often be modeled in organisms such as mouse and zebrafish to validate causation and understand developmental etiologies. Enhancers, however, are much less conserved across vertebrates and thus candidate non-coding mutations are more difficult to model. A few deeply conserved enhancers have been identified, such as a Pou3f3 enhancer associated with gill cover development in fishes and closure of embryonic neck cavities in mammals (Barske et al., 2020). For non-conserved enhancers, normal and patient-associated versions can be tested for differential activity in model organisms (Liu et al., 2020). Recently, the chromatin landscape of human embryonic craniofacial tissues has been analyzed, with putative enhancers correlated to craniofacial GWAS hits (Wilderman et al., 2018). A better understanding of craniofacial enhancer logic will be needed to predict which non-coding polymorphisms may be causative.
Variation in human facial shape and evolution of facial structure are likely driven primarily by changes in gene regulation. Integration of human facial variation GWAS (Shaffer et al., 2016; White et al., 2021) with single-cell craniofacial studies will help predict causative protein and non-coding polymorphisms. To date, single-cell studies of craniofacial development have been largely limited to zebrafish, mouse and human. Given that cell populations can be deconvolved by marker expression in single-cell datasets without the need for transgenic labels, it is now possible to perform single-cell analysis of craniofacial tissues from nearly any vertebrate. Comparative single-cell analyses should help inform how changes in gene expression and enhancer use correlate with evolution of facial form, as well as the extent to which cell types emerge or disappear during evolution. Single-cell approaches will complement exciting ongoing genomics analyses of avian beak shape (Smith et al., 2022), jaw length in Beloniformes fishes (Daane et al., 2021), and facial differences between apes, and archaic and modern humans (Gokhman et al., 2020; Prescott et al., 2015).
Finally, the application of single-cell technology to craniofacial regeneration would help reveal how resident progenitors and lineage plasticity contribute to repair. Single-cell analysis of the injured murine temporomandibular joint disc revealed potential progenitors for regeneration of the fibroblast but not chondrocyte components (Bi et al., 2023). In zebrafish, single-cell sequencing revealed a potential perichondral progenitor involved in regeneration of the jaw joint (Smeeton et al., 2021) and dedifferentiation of ligamentocytes during jaw ligament regeneration (Anderson et al., 2023). It will also be interesting to analyze whether regeneration-specific enhancers (Kang et al., 2016; Wang et al., 2020) function in craniofacial tissues, and whether, as for the heart (Yan et al., 2023), regeneration-specific enhancers can be leveraged to promote repair of mammalian craniofacial tissues. Comparative single-cell approaches of regenerative versus non-regenerative species will ultimately inform how differences in cell types, gene expression and enhancer usage underlie the ability of some vertebrates to robustly regenerate craniofacial tissues.
Footnotes
Funding
The authors’ research is funded by the National Institute of Dental and Craniofacial Research (NIH R35 DE027550) to J.G.C. and a California Institute for Regenerative Medicine EDUC4 Training Grant to K.-C.T. Deposited in PMC for release after 12 months.
References
- Aibar, S., González-Blas, C. B., Moerman, T., Huynh-Thu, V. A., Imrichova, H., Hulselmans, G., Rambow, F., Marine, J.-C., Geurts, P., Aerts, J.et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083-1086. 10.1038/nmeth.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghadeer, A., Hanson-Drury, S., Patni, A. P., Ehnes, D. D., Zhao, Y. T., Li, Z., Phal, A., Vincent, T., Lim, Y. C., O'Day, D.et al. (2023). Single-cell census of human tooth development enables generation of human enamel. Dev. Cell 10.1016/j.devcel.2023.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam, A., Nissen, R. M., Sun, Z., Swindell, E. C., Farrington, S. and Hopkins, N. (2004). Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 101, 12792-12797. 10.1073/pnas.0403929101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, T., Mo, J., Gagarin, E., Sherwood, D., Blumenkrantz, M., Mao, E., Leon, G., Chen, H.-J., Tseng, K.-C., Fabian, P.et al. (2023). Ligament injury in adult zebrafish triggers ECM remodeling and cell dedifferentiation for scar-free regeneration. npj Regen. Med. 8, 51. 10.1038/s41536-023-00329-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, C. D., Gerlach, D., Stelzer, C., Boryń, L. M., Rath, M. and Stark, A. (2013). Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339, 1074-1077. 10.1126/science.1232542 [DOI] [PubMed] [Google Scholar]
- Askary, A., Xu, P., Barske, L., Bay, M., Bump, P., Balczerski, B., Bonaguidi, M. A. and Crump, J. G. (2017). Genome-wide analysis of facial skeletal regionalization in zebrafish. Development 144, 2994-3005. 10.1242/dev.151712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askary, A., Sanchez-Guardado, L., Linton, J. M., Chadly, D. M., Budde, M. W., Cai, L., Lois, C. and Elowitz, M. B. (2020). In situ readout of DNA barcodes and single base edits facilitated by in vitro transcription. Nat. Biotechnol. 38, 66-75. 10.1038/s41587-019-0299-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balic, A. and Thesleff, I. (2015). Tissue interactions regulating tooth development and renewal. Curr. Top. Dev. Biol. 115, 157-186. 10.1016/bs.ctdb.2015.07.006 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay, A., Kubilus, J. K., Crochiere, M. L., Linsenmayer, T. F. and Tabin, C. J. (2008). Identification of unique molecular subdomains in the perichondrium and periosteum and their role in regulating gene expression in the underlying chondrocytes. Dev. Biol. 321, 162-174. 10.1016/j.ydbio.2008.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannier-Helaouet, M., Post, Y., Korving, J., Trani Bustos, M., Gehart, H., Begthel, H., Bar-Ephraim, Y. E., van der Vaart, J., Kalmann, R., Imhoff, S. M.et al. (2021). Exploring the human lacrimal gland using organoids and single-cell sequencing. Cell Stem Cell 28, 1221-1232.e1227. 10.1016/j.stem.2021.02.024 [DOI] [PubMed] [Google Scholar]
- Barske, L., Askary, A., Zuniga, E., Balczerski, B., Bump, P., Nichols, J. T. and Crump, J. G. (2016). Competition between Jagged-Notch and endothelin1 signaling selectively restricts cartilage formation in the zebrafish upper face. PLoS Genet. 12, e1005967. 10.1371/journal.pgen.1005967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barske, L., Fabian, P., Hirschberger, C., Jandzik, D., Square, T., Xu, P., Nelson, N., Yu, H. V., Medeiros, D. M., Gillis, J. A.et al. (2020). Evolution of vertebrate gill covers via shifts in an ancient Pou3f3 enhancer. Proc. Natl. Acad. Sci. USA 117, 24876-24884. 10.1073/pnas.2011531117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen, V., Lange, M., Peidli, S., Wolf, F. A. and Theis, F. J. (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408-1414. 10.1038/s41587-020-0591-3 [DOI] [PubMed] [Google Scholar]
- Beverdam, A., Merlo, G. R., Paleari, L., Mantero, S., Genova, F., Barbieri, O., Janvier, P. and Levi, G. (2002). Jaw transformation with gain of symmetry after Dlx5/Dlx6 inactivation: mirror of the past? Genesis 34, 221-227. 10.1002/gene.10156 [DOI] [PubMed] [Google Scholar]
- Bi, R., Yin, Q., Li, H., Yang, X., Wang, Y., Li, Q., Fang, H., Li, P., Lyu, P., Fan, Y.et al. (2023). A single-cell transcriptional atlas reveals resident progenitor cell niche functions in TMJ disc development and injury. Nat. Commun. 14, 830. 10.1038/s41467-023-36406-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bildsoe, H., Loebel, D. A. F., Jones, V. J., Chen, Y.-T., Behringer, R. R. and Tam, P. P. L. (2009). Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev. Biol. 331, 176-188. 10.1016/j.ydbio.2009.04.034 [DOI] [PubMed] [Google Scholar]
- Blentic, A., Tandon, P., Payton, S., Walshe, J., Carney, T., Kelsh, R. N., Mason, I. and Graham, A. (2008). The emergence of ectomesenchyme. Dev. Dyn. 237, 592-601. 10.1002/dvdy.21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist, R. F., Fowler, T. E., An, Z., Yu, T. Y., Abdilleh, K., Fraser, G. J., Sharpe, P. T. and Streelman, J. T. (2019). Developmental plasticity of epithelial stem cells in tooth and taste bud renewal. Proc. Natl. Acad. Sci. USA 116, 17858-17866. 10.1073/pnas.1821202116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booeshaghi, A. S., Gao, F. and Pachter, L. (2023). Assessing the multimodal tradeoff. bioRxiv 10.1101/2021.12.08.471788 [DOI] [Google Scholar]
- Brionne, A., Juanchich, A. and Hennequet-Antier, C. (2019). ViSEAGO: a Bioconductor package for clustering biological functions using Gene Ontology and semantic similarity. BioData Min. 12, 16. 10.1186/s13040-019-0204-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro, J. D., Wu, B., Litzenburger, U. M., Ruff, D., Gonzales, M. L., Snyder, M. P., Chang, H. Y. and Greenleaf, W. J. (2015). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486-490. 10.1038/nature14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdi, A. R. and Faist, K. (1967). Morphogenesis of the palate in normal human embryos with special emphasis on the mechanisms involved. Am. J. Anat. 120, 149-159. 10.1002/aja.1001200112 [DOI] [Google Scholar]
- Büttner, M., Miao, Z., Wolf, F. A., Teichmann, S. A. and Theis, F. J. (2019). A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods 16, 43-49. 10.1038/s41592-018-0254-1 [DOI] [PubMed] [Google Scholar]
- Caetano, A. J., Human Cell Atlas Oral and Craniofacial Bionetwork, Sequeira, I. and Byrd, K. M. (2022). A roadmap for the human oral and craniofacial cell atlas. J. Dent. Res. 101, 1274-1288. 10.1177/00220345221110768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J., Cusanovich, D. A., Ramani, V., Aghamirzaie, D., Pliner, H. A., Hill, A. J., Daza, R. M., McFaline-Figueroa, J. L., Packer, J. S., Christiansen, L.et al. (2018). Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380-1385. 10.1126/science.aau0730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J., Spielmann, M., Qiu, X., Huang, X., Ibrahim, D. M., Hill, A. J., Zhang, F., Mundlos, S., Christiansen, L., Steemers, F. J.et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496-502. 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, Y. and Maxson, R. E., Jr (2006). Recent advances in craniofacial morphogenesis. Dev. Dyn. 235, 2353-2375. 10.1002/dvdy.20833 [DOI] [PubMed] [Google Scholar]
- Chari, T. and Pachter, L. (2023). The specious art of single-cell genomics. PLoS Comput. Biol. 19, e1011288. 10.1371/journal.pcbi.1011288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Lin, W., Gan, J., Lu, W., Wang, M., Wang, X., Yi, J. and Zhao, Z. (2022). Transcriptomic mapping of human parotid gland at single-cell resolution. J. Dent. Res. 101, 972-982. 10.1177/00220345221076069 [DOI] [PubMed] [Google Scholar]
- Chen, H.-J., Barske, L., Talbot, J. C., Dinwoodie, O. M., Roberts, R. R., Farmer, D. T., Jimenez, C., Merrill, A. E., Tucker, A. S. and Crump, J. G. (2023). Nuclear receptor Nr5a2 promotes diverse connective tissue fates in the jaw. Dev. Cell 58, 461-473.e467. 10.1016/j.devcel.2023.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, C. P., Collazo, A., Trinh, L. A., Pan, L., Moens, C. B. and Crump, J. G. (2013). Wnt-dependent epithelial transitions drive pharyngeal pouch formation. Dev. Cell 24, 296-309. 10.1016/j.devcel.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collazo, A., Bronner-Fraser, M. and Fraser, S. E. (1993). Vital dye labelling of Xenopus laevis trunk neural crest reveals multipotency and novel pathways of migration. Development 118, 363-376. 10.1242/dev.118.2.363 [DOI] [PubMed] [Google Scholar]
- Colnot, C., Lu, C., Hu, D. and Helms, J. A. (2004). Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev. Biol. 269, 55-69. 10.1016/j.ydbio.2004.01.011 [DOI] [PubMed] [Google Scholar]
- Compagnucci, C., Martinus, K., Griffin, J. and Depew, M. J. (2021). Programmed cell death not as sledgehammer but as chisel: apoptosis in normal and abnormal craniofacial patterning and development. Front. Cell Dev. Biol. 9, 717404. 10.3389/fcell.2021.717404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coskun, A. F. and Cai, L. (2016). Dense transcript profiling in single cells by image correlation decoding. Nat. Methods 13, 657-660. 10.1038/nmeth.3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daane, J. M., Blum, N., Lanni, J., Boldt, H., Iovine, M. K., Higdon, C. W., Johnson, S. L., Lovejoy, N. R. and Harris, M. P. (2021). Modulation of bioelectric cues in the evolution of flying fishes. Curr. Biol. 31, 5052-5061.e5058. 10.1016/j.cub.2021.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, A. and Crump, J. G. (2012). Bmps and id2a act upstream of Twist1 to restrict ectomesenchyme potential of the cranial neural crest. PLoS Genet. 8, e1002710. 10.1371/journal.pgen.1002710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cuadra-Blanco, C., Peces-Peña, M. D. and Mérida-Velasco, J. R. (2003). Morphogenesis of the human lacrimal gland. J. Anat. 203, 531-536. 10.1046/j.1469-7580.2003.00233.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rop, F. V., Hulselmans, G., Flerin, C., Soler-Vila, P., Rafels, A., Christiaens, V., González-Blas, C. B., Marchese, D., Caratù, G., Poovathingal, S.et al. (2023). Systematic benchmarking of single-cell ATAC-sequencing protocols. Nat. Biotechnol. 10.1038/s41587-023-01881-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath, S., Yallowitz, A. R., McCormick, J., Lalani, S., Zhang, T., Xu, R., Li, N., Liu, Y., Yang, Y. S., Eiseman, M.et al. (2018). Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature 562, 133-139. 10.1038/s41586-018-0554-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew, M. J., Lufkin, T. and Rubenstein, J. L. R. (2002). Specification of jaw subdivisions by Dlx genes. Science 298, 381-385. 10.1126/science.1075703 [DOI] [PubMed] [Google Scholar]
- Ducy, P., Zhang, R., Geoffroy, V., Ridall, A. L. and Karsenty, G. (1997). Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747-754. 10.1016/S0092-8674(00)80257-3 [DOI] [PubMed] [Google Scholar]
- Efremova, M., Vento-Tormo, M., Teichmann, S. A. and Vento-Tormo, R. (2020). CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 15, 1484-1506. 10.1038/s41596-020-0292-x [DOI] [PubMed] [Google Scholar]
- Ehnes, D. D., Alghadeer, A., Hanson-Drury, S., Zhao, Y. T., Tilmes, G., Mathieu, J. and Ruohola-Baker, H. (2022). Sci-seq of human fetal salivary tissue introduces human transcriptional paradigms and a novel cell population. Front. Dent. Med. 3, 887057. 10.3389/fdmed.2022.887057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian, P. and Crump, J. G. (2022). Reassessing the embryonic origin and potential of craniofacial ectomesenchyme. Semin. Cell Dev. Biol. 138, 45-53. 10.1016/j.semcdb.2022.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian, P., Tseng, K.-C., Smeeton, J., Lancman, J. J., Dong, P. D. S., Cerny, R. and Crump, J. G. (2020). Lineage analysis reveals an endodermal contribution to the vertebrate pituitary. Science 370, 463-467. 10.1126/science.aba4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian, P., Tseng, K.-C., Thiruppathy, M., Arata, C., Chen, H.-J., Smeeton, J., Nelson, N. and Crump, J. G. (2022). Lifelong single-cell profiling of cranial neural crest diversification in zebrafish. Nat. Commun. 13, 13. 10.1038/s41467-021-27594-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J., Salathia, N., Liu, R., Kaeser, G. E., Yung, Y. C., Herman, J. L., Kaper, F., Fan, J.-B., Zhang, K., Chun, J.et al. (2016). Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 13, 241-244. 10.1038/nmeth.3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, R., Preissl, S., Li, Y., Hou, X., Lucero, J., Wang, X., Motamedi, A., Shiau, A. K., Zhou, X., Xie, F.et al. (2021). Comprehensive analysis of single cell ATAC-seq data with SnapATAC. Nat. Commun. 12, 1337. 10.1038/s41467-021-21583-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, D. T., Nathan, S., Finley, J. K., Shengyang Yu, K., Emmerson, E., Byrnes, L. E., Sneddon, J. B., McManus, M. T., Tward, A. D. and Knox, S. M. (2017). Defining epithelial cell dynamics and lineage relationships in the developing lacrimal gland. Development 144, 2517-2528. 10.1242/dev.150789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, D. J. T., Mlcochova, H., Zhou, Y., Koelling, N., Wang, G., Ashley, N., Bugacov, H., Chen, H.-J., Parvez, R., Tseng, K.-C.et al. (2021). The developing mouse coronal suture at single-cell resolution. Nat. Commun. 12, 4797. 10.1038/s41467-021-24917-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell, J. A., Wang, Y., Riesenfeld, S. J., Shekhar, K., Regev, A. and Schier, A. F. (2018). Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131. 10.1126/science.aar3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure, L., Soldatov, R., Kharchenko, P. V. and Adameyko, I. (2023). scFates: a scalable python package for advanced pseudotime and bifurcation analysis from single-cell data. Bioinformatics 39, btac746. 10.1093/bioinformatics/btac746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J., Han, X., Yuan, Y., Cho, C. K., Janečková, E., Guo, T., Pareek, S., Rahman, M. S., Zheng, B., Bi, J.et al. (2022). TGF-beta signaling and Creb5 cooperatively regulate Fgf18 to control pharyngeal muscle development. eLife 11, e80405. 10.7554/eLife.80405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieda, K. L., Linton, J. M., Hormoz, S., Choi, J., Chow, K.-H. K., Singer, Z. S., Budde, M. W., Elowitz, M. B. and Cai, L. (2017). Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107-111. 10.1038/nature20777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawel, D. R., Serra-Musach, J., Lilja, S., Aagesen, J., Arenas, A., Asking, B., Bengnér, M., Bjorkander, J., Biggs, S., Ernerudh, J.et al. (2019). A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Med. 11, 47. 10.1186/s13073-019-0657-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhman, D., Mishol, N., de Manuel, M., de Juan, D., Shuqrun, J., Meshorer, E., Marques-Bonet, T., Rak, Y. and Carmel, L. (2020). Reconstructing denisovan anatomy using DNA methylation maps. Cell 180, 601. 10.1016/j.cell.2020.01.020 [DOI] [PubMed] [Google Scholar]
- Gorin, G., Fang, M., Chari, T. and Pachter, L. (2022). RNA velocity unraveled. PLoS Comput. Biol. 18, e1010492. 10.1371/journal.pcbi.1010492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, A., Okabe, M. and Quinlan, R. (2005). The role of the endoderm in the development and evolution of the pharyngeal arches. J. Anat. 207, 479-487. 10.1111/j.1469-7580.2005.00472.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi, A. and Tajbakhsh, S. (2021). Diversity in cranial muscles: origins and developmental programs. Curr. Opin. Cell Biol. 73, 110-116. 10.1016/j.ceb.2021.06.005 [DOI] [PubMed] [Google Scholar]
- Grimaldi, A., Comai, G., Mella, S. and Tajbakhsh, S. (2022). Identification of bipotent progenitors that give rise to myogenic and connective tissues in mouse. eLife 11, e70235. 10.7554/eLife.70235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobstein, C. (1953). Inductive epitheliomesenchymal interaction in cultured organ rudiments of the mouse. Science 118, 52-55. 10.1126/science.118.3054.52 [DOI] [PubMed] [Google Scholar]
- Hammond, N. L. and Dixon, M. J. (2022). Revisiting the embryogenesis of lip and palate development. Oral Dis. 28, 1306-1326. 10.1111/odi.14174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, J., Mayo, J., Xu, X., Li, J., Bringas, P., Jr, Maas, R. L., Rubenstein, J. L. R. and Chai, Y. (2009). Indirect modulation of Shh signaling by Dlx5 affects the oral-nasal patterning of palate and rescues cleft palate in Msx1-null mice. Development 136, 4225-4233. 10.1242/dev.036723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, X., Feng, J., Guo, T., Loh, Y.-H. E., Yuan, Y., Ho, T.-V., Cho, C. K., Li, J., Jing, J., Janeckova, E.et al. (2021). Runx2-Twist1 interaction coordinates cranial neural crest guidance of soft palate myogenesis. eLife 10, e62387. 10.7554/eLife.62387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y., Hao, S., Andersen-Nissen, E., Mauck, W. M., Zheng, S., Butler, A., Lee, M. J., Wilk, A. J., Darby, C., Zager, M.et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573-3587.e3529. 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, B. R., Aure, M. H., Kelly, M. C., Genomics and Computational Biology Core, Hoffman, M. P. and Chibly, A. M. (2020). Generation of a single-cell RNAseq atlas of murine salivary gland development. iScience 23, 101838. 10.1016/j.isci.2020.101838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, G., Gonzalez-Reiche, A. S., Lu, N., Zhou, X., Rivera, J., Kriti, D., Sebra, R., Williams, A. A., Donovan, M. J., Potter, S. S.et al. (2020). Integrated transcriptome and network analysis reveals spatiotemporal dynamics of calvarial suturogenesis. Cell Rep. 32, 107871. 10.1016/j.celrep.2020.107871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, G., Gonzalez-Reiche, A. S., Saturne, M., Motch Perrine, S. M., Zhou, X., Borges, A. C., Shewale, B., Richtsmeier, J. T., Zhang, B., van Bakel, H.et al. (2021). Single-cell analysis identifies a key role for Hhip in murine coronal suture development. Nat. Commun. 12, 7132. 10.1038/s41467-021-27402-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horeth, E., Oyelakin, A., Song, E.-A. C., Che, M., Bard, J., Min, S., Kiripolsky, J., Kramer, J. M., Sinha, S. and Romano, R.-A. (2021). Transcriptomic and single-cell analysis reveals regulatory networks and cellular heterogeneity in mouse primary Sjogren's syndrome salivary glands. Front. Immunol. 12, 729040. 10.3389/fimmu.2021.729040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horeth, E., Bard, J., Che, M., Wrynn, T., Song, E. A. C., Marzullo, B., Burke, M. S., Popat, S., Loree, T., Zemer, J.et al. (2023). High-resolution transcriptomic landscape of the human submandibular gland. J. Dent. Res. 102, 525-535. 10.1177/00220345221147908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N., Pérez, P., Kato, T., Mikami, Y., Okuda, K., Gilmore, R. C., Conde, C. D., Gasmi, B., Stein, S., Beach, M.et al. (2021). SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 27, 892-903. 10.1038/s41591-021-01296-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, P., Gulisano, M., Cook, M., Sham, M.-H., Faiella, A., Wilkinson, D., Boncinelli, E. and Krumlauf, R. (1991). A distinct Hox code for the branchial region of the vertebrate head. Nature 353, 861-864. 10.1038/353861a0 [DOI] [PubMed] [Google Scholar]
- Inoue, F. and Ahituv, N. (2015). Decoding enhancers using massively parallel reporter assays. Genomics 106, 159-164. 10.1016/j.ygeno.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesh, K. A., Dey, K. K., Montoro, D. T., Mohan, R., Gazal, S., Engreitz, J. M., Xavier, R. J., Price, A. L. and Regev, A. (2022). Identifying disease-critical cell types and cellular processes by integrating single-cell RNA-sequencing and human genetics. Nat. Genet. 54, 1479-1492. 10.1038/s41588-022-01187-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskoll, T., Zhou, Y. M., Chai, Y., Makarenkova, H. P., Collinson, J. M., West, J. D., Hajihosseini, M. K., Lee, J. and Melnick, M. (2002). Embryonic submandibular gland morphogenesis: stage-specific protein localization of FGFs, BMPs, Pax6 and Pax9 in normal mice and abnormal SMG phenotypes in FgfR2-IIIc(+/Delta), BMP7(−/−) and Pax6(−/−) mice. Cells Tissues Organs 170, 83-98. 10.1159/000046183 [DOI] [PubMed] [Google Scholar]
- Jeong, J., Li, X., McEvilly, R. J., Rosenfeld, M. G., Lufkin, T. and Rubenstein, J. L. R. (2008). Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development 135, 2905-2916. 10.1242/dev.019778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, P., Hu, R., Yan, F., Dai, Y. and Zhao, Z. (2022). scGWAS: landscape of trait-cell type associations by integrating single-cell transcriptomics-wide and genome-wide association studies. Genome Biol. 23, 220. 10.1186/s13059-022-02785-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, X., Iseki, S., Maxson, R. E., Sucov, H. M. and Morriss-Kay, G. M. (2002). Tissue origins and interactions in the mammalian skull vault. Dev. Biol. 241, 106-116. 10.1006/dbio.2001.0487 [DOI] [PubMed] [Google Scholar]
- Jin, S., Guerrero-Juarez, C. F., Zhang, L., Chang, I., Ramos, R., Kuan, C.-H., Myung, P., Plikus, M. V. and Nie, Q. (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088. 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, J., Feng, J., Yuan, Y., Guo, T., Lei, J., Pei, F., Ho, T.-V. and Chai, Y. (2022). Spatiotemporal single-cell regulatory atlas reveals neural crest lineage diversification and cellular function during tooth morphogenesis. Nat. Commun. 13, 4803. 10.1038/s41467-022-32490-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, M. C., Noden, D. M., Hazelton, R. D., Coulombre, J. L. and Coulombre, A. J. (1979). Origins of avian ocular and periocular tissues. Exp. Eye Res. 29, 27-43. 10.1016/0014-4835(79)90164-7 [DOI] [PubMed] [Google Scholar]
- Justice, C. M., Cuellar, A., Bala, K., Sabourin, J. A., Cunningham, M. L., Crawford, K., Phipps, J. M., Zhou, Y., Cilliers, D., Byren, J. C.et al. (2020). A genome-wide association study implicates the BMP7 locus as a risk factor for nonsyndromic metopic craniosynostosis. Hum. Genet. 139, 1077-1090. 10.1007/s00439-020-02157-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadouri, N., Givony, T., Nevo, S., Hey, J., Ben Dor, S., Damari, G., Dassa, B., Dobes, J., Weichenhan, D., Bähr, M.et al. (2022). Transcriptional regulation of the thymus master regulator Foxn1. Sci Immunol. 7, eabn8144. 10.1126/sciimmunol.abn8144 [DOI] [PubMed] [Google Scholar]
- Kague, E., Gallagher, M., Burke, S., Parsons, M., Franz-Odendaal, T. and Fisher, S. (2012). Skeletogenic fate of zebrafish cranial and trunk neural crest. PLoS ONE 7, e47394. 10.1371/journal.pone.0047394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimoto, K., Stringa, B., Hoffmann, C. M., Jindal, K., Solnica-Krezel, L. and Morris, S. A. (2023). Dissecting cell identity via network inference and in silico gene perturbation. Nature 614, 742-751. 10.1038/s41586-022-05688-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J., Hu, J., Karra, R., Dickson, A. L., Tornini, V. A., Nachtrab, G., Gemberling, M., Goldman, J. A., Black, B. L. and Poss, K. D. (2016). Modulation of tissue repair by regeneration enhancer elements. Nature 532, 201-206. 10.1038/nature17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartha, V. K., Duarte, F. M., Hu, Y., Ma, S., Chew, J. G., Lareau, C. A., Earl, A., Burkett, Z. D., Kohlway, A. S., Lebofsky, R.et al. (2022). Functional inference of gene regulation using single-cell multi-omics. Cell Genomics 2, 100166. 10.1016/j.xgen.2022.100166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyabu, D., Maier, W. and Sánchez-Villagra, M. R. (2012). Paleontological and developmental evidence resolve the homology and dual embryonic origin of a mammalian skull bone, the interparietal. Proc. Natl. Acad. Sci. USA 109, 14075-14080. 10.1073/pnas.1208693109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivanek, J., Soldatov, R. A., Kastriti, M. E., Chontorotzea, T., Herdina, A. N., Petersen, J., Szarowska, B., Landova, M., Matejova, V. K., Holla, L. I.et al. (2020). Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat. Commun. 11, 4816. 10.1038/s41467-020-18512-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno, G., Soldatov, R., Zeisel, A., Braun, E., Hochgerner, H., Petukhov, V., Lidschreiber, K., Kastriti, M. E., Lönnerberg, P., Furlan, A.et al. (2018). RNA velocity of single cells. Nature 560, 494-498. 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, M., Bergen, V., Klein, M., Setty, M., Reuter, B., Bakhti, M., Lickert, H., Ansari, M., Schniering, J., Schiller, H. B.et al. (2022). CellRank for directed single-cell fate mapping. Nat. Methods 19, 159-170. 10.1038/s41592-021-01346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Lievre, C. S. and Le Douarin, N. M. (1975). Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J. Embryol. Exp. Morphol. 34, 125-154. 10.1242/dev.34.1.125 [DOI] [PubMed] [Google Scholar]
- Lescroart, F., Hamou, W., Francou, A., Théveniau-Ruissy, M., Kelly, R. G. and Buckingham, M. (2015). Clonal analysis reveals a common origin between nonsomite-derived neck muscles and heart myocardium. Proc. Natl. Acad. Sci. USA 112, 1446-1451. 10.1073/pnas.1424538112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, E. J., Liu, H., Carlson, J. C., Shaffer, J. R., Feingold, E., Wehby, G., Laurie, C. A., Jain, D., Laurie, C. C., Doheny, K. F.et al. (2016). A genome-wide association study of nonsyndromic cleft palate identifies an etiologic missense variant in GRHL3. Am. J. Hum. Genet. 98, 744-754. 10.1016/j.ajhg.2016.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, E. J., Carlson, J. C., Shaffer, J. R., Butali, A., Buxó, C. J., Castilla, E. E., Christensen, K., Deleyiannis, F. W. B., Leigh Field, L., Hecht, J. T.et al. (2017). Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet. 136, 275-286. 10.1007/s00439-016-1754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Jones, K. L., Hooper, J. E. and Williams, T. (2019). The molecular anatomy of mammalian upper lip and primary palate fusion at single cell resolution. Development 146, dev174888. 10.1242/dev.174888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Wachsmuth, L. P., Xiao, S., Condie, B. G. and Manley, N. R. (2023). Foxn1 overexpression promotes thymic epithelial progenitor cell proliferation and mTEC maintenance, but does not prevent thymic involution. Development 150, dev200995. 10.1242/dev.200995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Li, C., Xu, J., Lan, Y., Liu, H., Li, X., Maire, P., Wang, X. and Jiang, R. (2019). Crucial and overlapping roles of Six1 and Six2 in craniofacial development. J. Dent. Res. 98, 572-579. 10.1177/0022034519835204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Duncan, K., Helverson, A., Kumari, P., Mumm, C., Xiao, Y., Carlson, J. C., Darbellay, F., Visel, A., Leslie, E.et al. (2020). Analysis of zebrafish periderm enhancers facilitates identification of a regulatory variant near human KRT8/18. eLife 9, e51325. 10.7554/eLife.51325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert, I. M. A., Abrams, S. R., Li, L., Eswarakumar, V. P., Sethi, A. J., Witt, R. L. and Hoffman, M. P. (2013). Combined KIT and FGFR2b signaling regulates epithelial progenitor expansion during organogenesis. Stem Cell Rep. 1, 604-619. 10.1016/j.stemcr.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magaletta, M. E., Lobo, M., Kernfeld, E. M., Aliee, H., Huey, J. D., Parsons, T. J., Theis, F. J. and Maehr, R. (2022). Integration of single-cell transcriptomes and chromatin landscapes reveals regulatory programs driving pharyngeal organ development. Nat. Commun. 13, 457. 10.1038/s41467-022-28067-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Madrigal, F. and Micheau, C. (1989). Histology of the major salivary glands. Am. J. Surg. Pathol. 13, 879-899. 10.1097/00000478-198910000-00008 [DOI] [PubMed] [Google Scholar]
- Maruyama, T., Jeong, J., Sheu, T.-J. and Hsu, W. (2016). Stem cells of the suture mesenchyme in craniofacial bone development, repair and regeneration. Nat. Commun. 7, 10526. 10.1038/ncomms10526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita, Y., Nagata, M., Kozloff, K. M., Welch, J. D., Mizuhashi, K., Tokavanich, N., Hallett, S. A., Link, D. C., Nagasawa, T., Ono, W.et al. (2020). A Wnt-mediated transformation of the bone marrow stromal cell identity orchestrates skeletal regeneration. Nat. Commun. 11, 332. 10.1038/s41467-019-14029-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, S., Salhotra, A., Shailendra, S., Tevlin, R., Ransom, R. C., Januszyk, M., Chan, C. K. F., Behr, B., Wan, D. C., Longaker, M. T.et al. (2021). Skeletal stem and progenitor cells maintain cranial suture patency and prevent craniosynostosis. Nat. Commun. 12, 4640. 10.1038/s41467-021-24801-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill, A. E., Bochukova, E. G., Brugger, S. M., Ishii, M., Pilz, D. T., Wall, S. A., Lyons, K. M., Wilkie, A. O. M. and Maxson, R. E., Jr (2006). Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum. Mol. Genet. 15, 1319-1328. 10.1093/hmg/ddl052 [DOI] [PubMed] [Google Scholar]
- Miao, Z., Deng, K., Wang, X. and Zhang, X. (2018). DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics 34, 3223-3224. 10.1093/bioinformatics/bty332 [DOI] [PubMed] [Google Scholar]
- Miller, C. T., Maves, L. and Kimmel, C. B. (2004). moz regulates Hox expression and pharyngeal segmental identity in zebrafish. Development 131, 2443-2461. 10.1242/dev.01134 [DOI] [PubMed] [Google Scholar]
- Millicovsky, G., Ambrose, L. J. H. and Johnston, M. C. (1982). Developmental alterations associated with spontaneous cleft lip and palate in CL/Fr mice. Am. J. Anat. 164, 29-44. 10.1002/aja.1001640104 [DOI] [PubMed] [Google Scholar]
- Minoux, M. and Rijli, F. M. (2010). Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 137, 2605-2621. 10.1242/dev.040048 [DOI] [PubMed] [Google Scholar]
- Mitchell, J. M., Sucharov, J., Pulvino, A. T., Brooks, E. P., Gillen, A. E. and Nichols, J. T. (2021). The alx3 gene shapes the zebrafish neurocranium by regulating frontonasal neural crest cell differentiation timing. Development 148, dev197483. 10.1242/dev.197483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongera, A., Singh, A. P., Levesque, M. P., Chen, Y.-Y., Konstantinidis, P. and Nüsslein-Volhard, C. (2013). Genetic lineage labeling in zebrafish uncovers novel neural crest contributions to the head, including gill pillar cells. Development 140, 916-925. 10.1242/dev.091066 [DOI] [PubMed] [Google Scholar]
- Moon, K. R., van Dijk, D., Wang, Z., Gigante, S., Burkhardt, D. B., Chen, W. S., Yim, K., Elzen, A. v. d., Hirn, M. J., Coifman, R. R.et al. (2019). Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol. 37, 1482-1492. 10.1038/s41587-019-0336-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, J. A., McLennan, R., Wolfe, L. A., Gogol, M. M., Meier, S., McKinney, M. C., Teddy, J. M., Holmes, L., Semerad, C. L., Box, A. C.et al. (2017). Single-cell transcriptome analysis of avian neural crest migration reveals signatures of invasion and molecular transitions. eLife 6, e28415. 10.7554/eLife.28415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, J. A., McLennan, R., Teddy, J. M., Scott, A. R., Kasemeier-Kulesa, J. C., Gogol, M. M. and Kulesa, P. M. (2021). Single-cell reconstruction with spatial context of migrating neural crest cells and their microenvironments during vertebrate head and neck formation. Development 148, dev199468. 10.1242/dev.199468 [DOI] [PubMed] [Google Scholar]
- Moskwa, N., Mahmood, A., Nelson, D. A., Altrieth, A. L., Forni, P. E. and Larsen, M. (2022). Single-cell RNA sequencing reveals PDFGRalpha+ stromal cell subpopulations that promote proacinar cell differentiation in embryonic salivary gland organoids. Development 149, dev200167. 10.1242/dev.200167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, S. A. (2011). Mouse resources for craniofacial research. Genesis 49, 190-199. 10.1002/dvg.20722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayar, S., Turner, J. D., Asam, S., Fennell, E., Pugh, M., Colfrancesco, S., Berardicurti, O., Smith, C. G., Flint, J., Doedosio, A.et al. (2022). A cellular and spatial map of salivary glands at single cell resolution reveals the functional basis of tertiary lymphoid structure formation in Sjogren's syndrome. bioRxiv 10.1101/2022.11.03.514908 [DOI] [Google Scholar]
- Nomaru, H., Liu, Y., De Bono, C., Righelli, D., Cirino, A., Wang, W., Song, H., Racedo, S. E., Dantas, A. G., Zhang, L.et al. (2021). Single cell multi-omic analysis identifies a Tbx1-dependent multilineage primed population in murine cardiopharyngeal mesoderm. Nat. Commun. 12, 6645. 10.1038/s41467-021-26966-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyelakin, A., Song, E. A. C., Min, S., Bard, J. E., Kann, J. V., Horeth, E., Smalley, K., Kramer, J. M., Sinha, S. and Romano, R. A. (2019). Transcriptomic and single-cell analysis of the murine parotid gland. J. Dent. Res. 98, 1539-1547. 10.1177/0022034519882355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozekin, Y. H., O'Rourke, R. and Bates, E. A. (2023). Single cell sequencing of the mouse anterior palate reveals mesenchymal heterogeneity. Dev. Dyn. 252, 713-727. 10.1002/dvdy.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski, T., Schilling, T. F., Brand, M., Jiang, Y.-J., Heisenberg, C.-P., Beuchle, D., Grandel, H., van Eeden, F. J., Furutani-Seiki, M., Granato, M.et al. (1996). Jaw and branchial arch mutants in zebrafish II: anterior arches and cartilage differentiation. Development 123, 345-356. 10.1242/dev.123.1.345 [DOI] [PubMed] [Google Scholar]
- Platt, J. B. (1893). Ectodermic origin of the cartilages of the head. Anat. Anz. 8, 506-509. [Google Scholar]
- Prescott, S. L., Srinivasan, R., Marchetto, M. C., Grishina, I., Narvaiza, I., Selleri, L., Gage, F. H., Swigut, T. and Wysocka, J. (2015). Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163, 68-83. 10.1016/j.cell.2015.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushel, I., Trainor, P. A. and Krumlauf, R. (2021). Specification of axial identity by Hoxa2 distinguishes between a phenotypic and molecular ground state in mouse cranial neural crest cells. bioRxiv 10.1101/2021.02.09.430457 [DOI] [Google Scholar]
- Qiu, X., Mao, Q., Tang, Y., Wang, L., Chawla, R., Pliner, H. A. and Trapnell, C. (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979-982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransick, A., Lindstrom, N. O., Liu, J., Zhu, Q., Guo, J.-J., Alvarado, G. F., Kim, A. D., Black, H. G., Kim, J. and McMahon, A. P. (2019). Single-cell profiling reveals sex, lineage, and regional diversity in the mouse kidney. Dev. Cell 51, 399-413.e397. 10.1016/j.devcel.2019.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson, R. J., Hammond, N. L., Coulombe, P. A., Saloranta, C., Nousiainen, H. O., Salonen, R., Berry, A., Hanley, N., Headon, D., Karikoski, R.et al. (2014). Periderm prevents pathological epithelial adhesions during embryogenesis. J. Clin. Invest. 124, 3891-3900. 10.1172/JCI71946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman, L. P., Goyal, Y., Jiang, C. L. and Raj, A. (2023). ClonoCluster: a method for using clonal origin to inform transcriptome clustering. Cell Genom 3, 100247. 10.1016/j.xgen.2022.100247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothova, M., Thompson, H., Lickert, H. and Tucker, A. S. (2012). Lineage tracing of the endoderm during oral development. Dev. Dyn. 241, 1183-1191. 10.1002/dvdy.23804 [DOI] [PubMed] [Google Scholar]
- Saelens, W., Cannoodt, R., Todorov, H. and Saeys, Y. (2019). A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547-554. 10.1038/s41587-019-0071-9 [DOI] [PubMed] [Google Scholar]
- Schilling, T. F., Piotrowski, T., Grandel, H., Brand, M., Heisenberg, C.-P., Jiang, Y.-J., Beuchle, D., Hammerschmidt, M., Kane, D. A., Mullins, M. C.et al. (1996). Jaw and branchial arch mutants in zebrafish I: branchial arches. Development 123, 329-344. 10.1242/dev.123.1.329 [DOI] [PubMed] [Google Scholar]
- Schlosser, G. (2017). From so simple a beginning - what amphioxus can teach us about placode evolution. Int. J. Dev. Biol. 61, 633-648. 10.1387/ijdb.170127gs [DOI] [PubMed] [Google Scholar]
- Seidel, K., Marangoni, P., Tang, C., Houshmand, B., Du, W., Maas, R. L., Murray, S., Oldham, M. C. and Klein, O. D. (2017). Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. eLife 6, e24712. 10.7554/eLife.24712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi, R., Martin, D. and Yamada, K. M. (2020). Single-cell RNA-seq identifies cell diversity in embryonic salivary glands. J. Dent. Res. 99, 69-78. 10.1177/0022034519883888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, J. R., Orlova, E., Lee, M. K., Leslie, E. J., Raffensperger, Z. D., Heike, C. L., Cunningham, M. L., Hecht, J. T., Kau, C. H., Nidey, N. L.et al. (2016). Genome-wide association study reveals multiple loci influencing normal human facial morphology. PLoS Genet. 12, e1006149. 10.1371/journal.pgen.1006149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F., Phan, A. V., Can Otterloo, E. and Cao, H. (2022). Transcriptional programs controlling lineages specification of mandibular epithelium during tooth initiation. bioRxiv. 10.1101/2022.11.26.518052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharir, A., Marangoni, P., Zilionis, R., Wan, M., Wald, T., Hu, J. K., Kawaguchi, K., Castillo-Azofeifa, D., Epstein, L., Harrington, K.et al. (2019). A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nat. Cell Biol. 21, 1102-1112. 10.1038/s41556-019-0378-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewert, A., Reiz, B., Krug, C., Heggemann, J., Mangold, E., Dickten, H. and Ludwig, K. U. (2023). Analysis of candidate genes for cleft lip±cleft palate using murine single-cell expression data. Front. Cell Dev. Biol. 11, 1091666. 10.3389/fcell.2023.1091666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleight, V. A. and Gillis, J. A. (2020). Embryonic origin and serial homology of gill arches and paired fins in the skate, Leucoraja erinacea. eLife 9, e60635. 10.7554/eLife.60635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeeton, J., Natarajan, N., Anderson, T., Tseng, K.-C., Fabian, P. and Crump, J. G. (2021). Regeneration of jaw joint cartilage in adult zebrafish. Front. Cell Dev. Biol. 9, 777787. 10.3389/fcell.2021.777787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. S., Chu, D., Qu, T., Aggleton, J. A. and Schneider, R. A. (2022). Species-specific sensitivity to TGFbeta signaling and changes to the Mmp13 promoter underlie avian jaw development and evolution. eLife 11, e66005. 10.7554/eLife.66005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov, R., Kaucka, M., Kastriti, M. E., Petersen, J., Chontorotzea, T., Englmaier, L., Akkuratova, N., Yang, Y., Haring, M., Dyachuk, V.et al. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536. 10.1126/science.aas9536 [DOI] [PubMed] [Google Scholar]
- Song, E.-A. C., Min, S., Oyelakin, A., Smalley, K., Bard, J. E., Liao, L., Xu, J. and Romano, R.-A. (2018). Genetic and scRNA-seq analysis reveals distinct cell populations that contribute to salivary gland development and maintenance. Sci. Rep. 8, 14043. 10.1038/s41598-018-32343-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, M., Yuan, X., Racioppi, C., Leslie, M., Stutt, N., Aleksandrova, A., Christiaen, L., Wilson, M. D. and Scott, I. C. (2022). GATA4/5/6 family transcription factors are conserved determinants of cardiac versus pharyngeal mesoderm fate. Sci. Adv. 8, eabg0834. 10.1126/sciadv.abg0834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhl, P. L., Salmén, F., Vickovic, S., Lundmark, A., Navarro, J. F., Magnusson, J., Giacomello, S., Asp, M., Westholm, J. O., Huss, M.et al. (2016). Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78-82. 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- Stolper, J., Ambrosio, E. M., Danciu, D.-P., Buono, L., Elliott, D. A., Naruse, K., Martinez-Morales, J. R., Marciniak-Czochra, A. and Centanin, L. (2019). Stem cell topography splits growth and homeostatic functions in the fish gill. eLife 8, e43747. 10.7554/eLife.43747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. and Satija, R. (2021). Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333-1341. 10.1038/s41592-021-01282-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Lin, Y., Ha, N., Zhang, J., Wang, W., Wang, X. and Bian, Q. (2023). Single-cell RNA-Seq reveals transcriptional regulatory networks directing the development of mouse maxillary prominence. J. Genet. Genomics. 10.1016/j.jgg.2023.02.008 [DOI] [PubMed] [Google Scholar]
- Talbot, J. C., Johnson, S. L. and Kimmel, C. B. (2010). hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 137, 2507-2517. 10.1242/dev.049700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W. and Bronner, M. E. (2020). Neural crest lineage analysis: from past to future trajectory. Development 147, dev193193. 10.1242/dev.193193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarashansky, A. J., Musser, J. M., Khariton, M., Li, P., Arendt, D., Quake, S. R. and Wang, B. (2021). Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife 10, e66747. 10.7554/eLife.66747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarakis, D., Cang, Z., Wu, X., Sharma, P. P., Karikomi, M., MacLean, A. L., Nie, Q. and Schilling, T. F. (2021). Single-cell transcriptomic analysis of zebrafish cranial neural crest reveals spatiotemporal regulation of lineage decisions during development. Cell Rep. 37, 110140. 10.1016/j.celrep.2021.110140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Berge, D., Brouwer, A., Korving, J., Martin, J. F. and Meijlink, F. (1998). Prx1 and Prx2 in skeletogenesis: roles in the craniofacial region, inner ear and limbs. Development 125, 3831-3842. 10.1242/dev.125.19.3831 [DOI] [PubMed] [Google Scholar]
- Teng, C. S., Cavin, L., Maxson, R. E., Jr, Sanchez-Villagra, M. R. and Crump, J. G. (2019). Resolving homology in the face of shifting germ layer origins: Lessons from a major skull vault boundary. eLife 8, e52814. 10.7554/eLife.52814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, H., Ohazama, A., Sharpe, P. T. and Tucker, A. S. (2012). The origin of the stapes and relationship to the otic capsule and oval window. Dev. Dyn. 241, 1396-1404. 10.1002/dvdy.23831 [DOI] [PubMed] [Google Scholar]
- Ting, M.-C., Wu, N. L., Roybal, P. G., Sun, J., Liu, L., Yen, Y. and Maxson, R. E., Jr (2009). EphA4 as an effector of Twist1 in the guidance of osteogenic precursor cells during calvarial bone growth and in craniosynostosis. Development 136, 855-864. 10.1242/dev.028605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower, R. J., Li, Z., Cheng, Y.-H., Wang, X.-W., Rajbhandari, L., Zhang, Q., Negri, S., Uytingco, C. R., Venkatesan, A., Zhou, F.-Q.et al. (2021). Spatial transcriptomics reveals a role for sensory nerves in preserving cranial suture patency through modulation of BMP/TGF-beta signaling. Proc. Natl. Acad. Sci. USA 118, e2103087118. 10.1073/pnas.2103087118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, A. S. (2007). Salivary gland development. Semin. Cell Dev. Biol. 18, 237-244. 10.1016/j.semcdb.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Twigg, S. R. F. and Wilkie, A. O. M. (2015). A genetic-pathophysiological framework for craniosynostosis. Am. J. Hum. Genet. 97, 359-377. 10.1016/j.ajhg.2015.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Otterloo, E., Williams, T. and Artinger, K. B. (2016). The old and new face of craniofacial research: How animal models inform human craniofacial genetic and clinical data. Dev. Biol. 415, 171-187. 10.1016/j.ydbio.2016.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D. E., Weinreb, C., Collins, Z. M., Briggs, J. A., Megason, S. G. and Klein, A. M. (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981-987. 10.1126/science.aar4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Hu, C.-K., Zeng, A., Alegre, D., Hu, D., Gotting, K., Ortega Granillo, A., Wang, Y., Robb, S., Schnittker, R.et al. (2020). Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 369, eaaz3090. 10.1126/science.aaz3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb, C., Wolock, S. and Klein, A. M. (2017). SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246-1248. 10.1093/bioinformatics/btx792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb, C., Wolock, S., Tusi, B. K., Socolovsky, M. and Klein, A. M. (2018). Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl. Acad. Sci. USA 115, E2467-E2476. 10.1073/pnas.1714723115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, J. A., Yoshida, H., Robinson, V., Nishikawa, S., Fraser, S. T. and Nishikawa, S. (2004). Neural crest and the origin of ectomesenchyme: neural fold heterogeneity suggests an alternative hypothesis. Dev. Dyn. 229, 118-130. 10.1002/dvdy.10478 [DOI] [PubMed] [Google Scholar]
- White, J. D., Indencleef, K., Naqvi, S., Eller, R. J., Hoskens, H., Roosenboom, J., Lee, M. K., Li, J., Mohammed, J., Richmond, S.et al. (2021). Insights into the genetic architecture of the human face. Nat. Genet. 53, 45-53. 10.1038/s41588-020-00741-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman, A., VanOudenhove, J., Kron, J., Noonan, J. P. and Cotney, J. (2018). High-resolution epigenomic atlas of human embryonic craniofacial development. Cell Rep. 23, 1581-1597. 10.1016/j.celrep.2018.03.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. M., Candido-Ferreira, I., Repapi, E., Gavriouchkina, D., Senanayake, U., Ling, I. T. C., Telenius, J., Taylor, S., Hughes, J. and Sauka-Spengler, T. (2019). Reconstruction of the global neural crest gene regulatory network in vivo. Dev. Cell 51, 255-276.e257. 10.1016/j.devcel.2019.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, F. A., Angerer, P. and Theis, F. J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. 10.1186/s13059-017-1382-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, F. A., Hamey, F. K., Plass, M., Solana, J., Dahlin, J. S., Göttgens, B., Rajewsky, N., Simon, L. and Theis, F. J. (2019). PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59. 10.1186/s13059-019-1663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, B. and Yanai, I. (2019). A periodic table of cell types. Development 146, dev169854. 10.1242/dev.169854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, C., Fan, J., Emanuel, G., Hao, J. and Zhuang, X. (2019). Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. Proc. Natl. Acad. Sci. USA 116, 19490-19499. 10.1073/pnas.1912459116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Liu, H., Lan, Y., Adam, M., Clouthier, D. E., Potter, S. and Jiang, R. (2019). Hedgehog signaling patterns the oral-aboral axis of the mandibular arch. eLife 8, e40315. 10.7554/eLife.40315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R., Liu, Y., Zhou, Y., Lin, W., Yuan, Q., Zhou, X. and Yang, Y. (2022). Gnas loss causes chondrocyte fate conversion in cranial suture formation. J. Dent. Res. 101, 931-941. 10.1177/00220345221075215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, R., Cigliola, V., Oonk, K. A., Petrover, Z., DeLuca, S., Wolfson, D. W., Vekstein, A., Mendiola, M. A., Devlin, G., Bishawi, M.et al. (2023). An enhancer-based gene-therapy strategy for spatiotemporal control of cargoes during tissue repair. Cell Stem Cell 30, 96-111.e116. 10.1016/j.stem.2022.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankee, T. N., Oh, S., Winchester, E. W., Wilderman, A., Robingson, K., Gordon, T., Rosenfeld, J. A., VanOudenhove, J., Scott , D., et al. (2023). Integrative analysis of transcriptomics in human craniofacial development reveals novel candidate disease genes. Nat. Commun. 14, 4623. 10.1038/s41467-023-40363-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Q., Bhojwani, A. and Hu, J. K. (2022). Understanding the development of oral epithelial organs through single cell transcriptomic analysis. Development 149, dev200539. 10.1242/dev.200539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., Gu, S., Alappat, S., Song, Y., Yan, M., Zhang, X., Zhang, G., Jiang, Y., Zhang, Z., Zhang, Y.et al. (2005). Shox2-deficient mice exhibit a rare type of incomplete clefting of the secondary palate. Development 132, 4397-4406. 10.1242/dev.02013 [DOI] [PubMed] [Google Scholar]
- Yu, M., Ma, L., Yuan, Y., Ye, X., Montagne, A., He, J., Ho, T.-V., Wu, Y., Zhao, Z., Sta Maria, N.et al. (2021). Cranial suture regeneration mitigates skull and neurocognitive defects in craniosynostosis. Cell 184, 243-256.e218. 10.1016/j.cell.2020.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y., Loh, Y.-E., Han, X., Feng, J., Ho, T.-V., He, J., Jing, J., Groff, K., Wu, A. and Chai, Y. (2020). Spatiotemporal cellular movement and fate decisions during first pharyngeal arch morphogenesis. Sci. Adv. 6, eabb0119. 10.1126/sciadv.abb0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M. J., Hou, K., Dey, K. K., Sakaue, S., Jagadeesh, K. A., Weinand, K., Taychameekiatchai, A., Rao, P., Pisco, A. O., Zou, J.et al. (2022). Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data. Nat. Genet. 54, 1572-1580. 10.1038/s41588-022-01167-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., Feng, J., Ho, T.-V., Grimes, W., Urata, M. and Chai, Y. (2015). The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol. 17, 386-396. 10.1038/ncb3139 [DOI] [PMC free article] [PubMed] [Google Scholar]