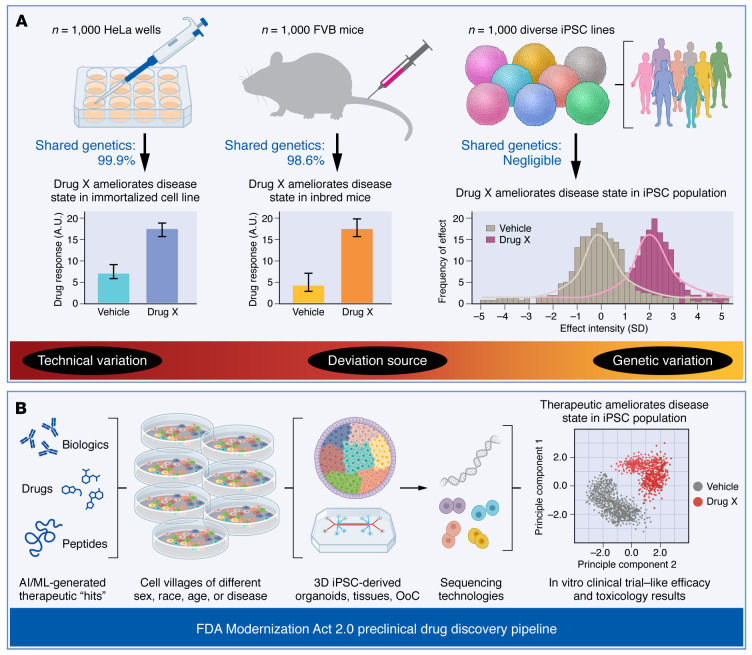
Figure 1. Lack of genetic diversity and pharmacogenomic differences between model animals and humans lead to the high termination rate of phase I and II clinical trials.
(A) Immortalized cell lines such as HeLa cells and inbred rodent models are commonly used as the gold standard of preclinical validation due to their ease of use. However, tissue cultures use immortalized cell lines with 99.9% shared genetics on average and rodent experiments use inbred strains with 98.6% shared genetics on average. Data from technical replicates of therapeutic agents can have positive results with low standard deviations that may be misleading. In contrast, testing therapeutic agents on patient-specific iPSCs more accurately represents the full genetic and pharmacogenomic diversity of human populations. Here, experimental results would reflect the effects of therapeutic agents in responders and nonresponders across a large cohort of individuals (rather than in a single immortalized cell line or a chosen rodent strain), hence providing more reliable safety and efficacy data prior to proceeding to clinical trials. (B) Innovative techniques for cultivating multiple iPSC lines together and segregating their transcriptomic and genetic signals have given rise to the concept of “cell villages.” These pooled populations are then differentiated into various cell types and incorporated into 3D models such as organoids or organs-on-chips, closely mimicking the human in vivo environment. These cell village tissues can then be subjected to therapeutic agents identified from AI/ML models, and the application of single-cell technologies allows for elucidation of each cell line’s distinct gene expression response to the therapeutic intervention. This transformative approach of clinical-trial-in-a-dish holds immense potential for enhancing our understanding of drug safety and efficacy, ultimately increasing the likelihood of therapeutic success in early clinical trials.