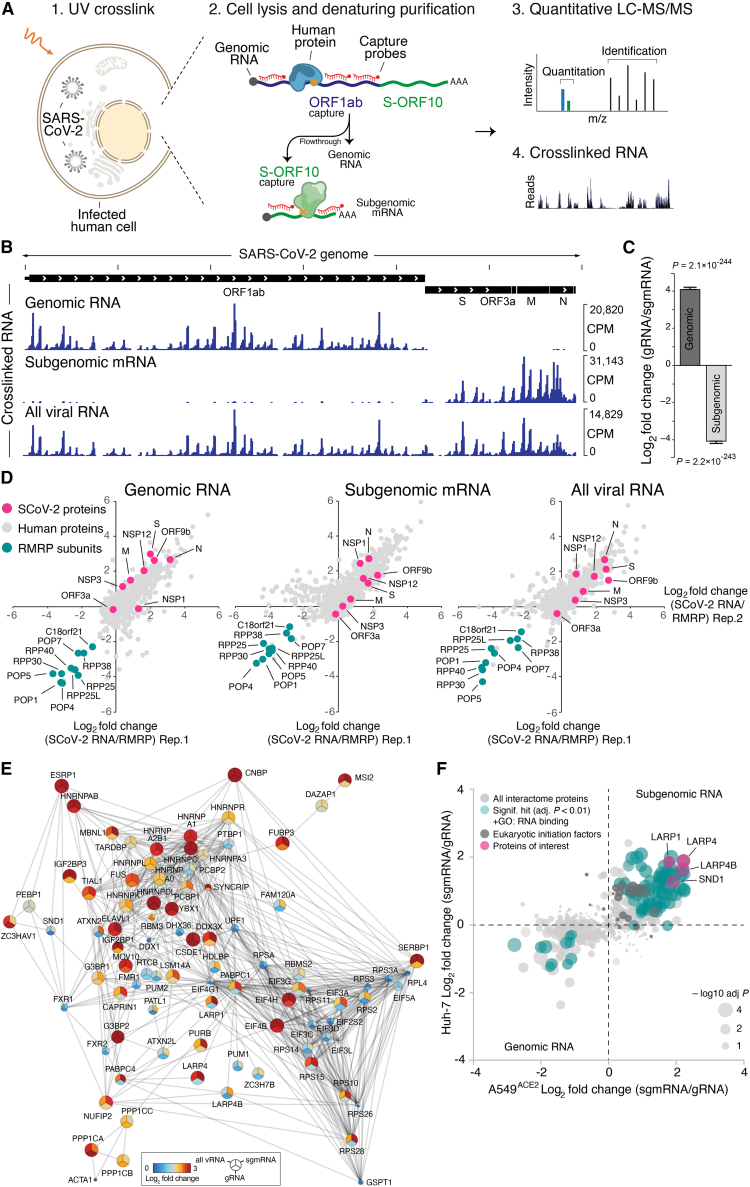
Figure 1.
The SARS-CoV-2 RNA-protein interactome at subgenome resolution
(A) Outline of RAP-MS workflow to identify proteins bound to SARS-CoV-2 genomic RNA (gRNA) and subgenomic mRNA (sgmRNA).
(B) Sequencing of RNA crosslinked to proteins purified with RAP-MS.
(C) Enrichment of reads mapping to SARS-CoV-2 genomic and subgenomic RNA in gRNA relative to sgmRNA purifications ± SE. p values determined by Wald test.
(D) Quantification of SARS-CoV-2 RNA interacting proteins relative to RMRP interacting proteins in antisense purifications of indicated RNA species. Log2 fold changes from two biological replicates in Huh-7 cells are shown. Gray, all detected proteins; magenta, SARS-CoV-2 proteins; teal, RMRP components.
(E) Protein-protein association network for consensus SARS-CoV-2 RNA interactome based on interactions in STRING v11.13 Coloring indicates RAP-MS enrichment in Huh-7 cells. Nodes are scaled to significance.
(F) Quantification of sgmRNA relative to gRNA interactomes. Average log2 fold changes in A549ACE2 cells (x axis) and Huh-7 cells (y axis) are shown. Circles are scaled to significance. Keratins and carboxylases were removed.
(E and F) Combined p values calculated using Fisher’s method for combined probability and adjusted using Benjamini-Hochberg procedure.