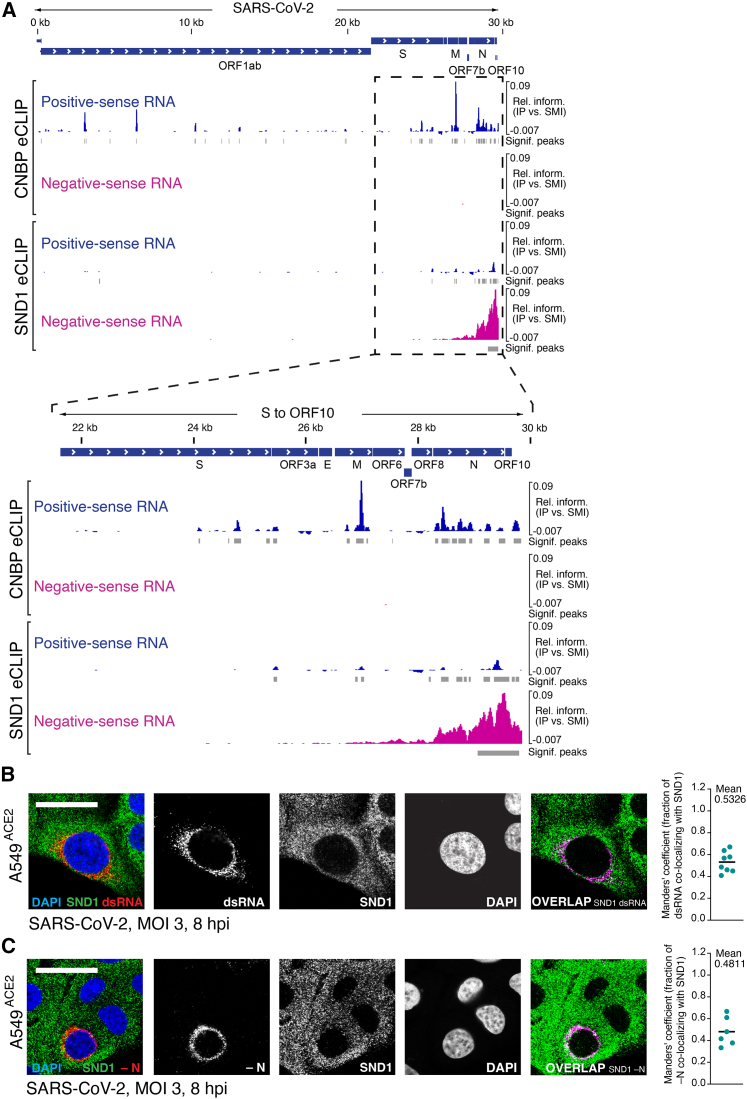
Figure 3.
SND1 binds negative-sense viral RNA
(A) Alignment of strand-separated eCLIP data for SND1 and CNBP8 to SARS-CoV-2 genome. Relative information in IP vs. size-matched input (SMI) is calculated at each position (STAR Methods) and displayed for positive-sense (blue) and negative-sense RNA (magenta). Peaks significantly enriched relative to SMI are indicated in gray. Lower panel displays zoom in to the S to ORF10 region. eCLIP was performed in Huh-7 cells at 24 hpi.
(B) IF staining of SND1 and dsRNA in SARS-CoV-2 infected A549ACE2 cells at 8 hpi (MOI = 3 PFU/cell). Representative images are shown. Overlap is quantified by Manders’ co-localization coefficient (n = 8 images). Uninfected cells shown in Figure S3A.
(C) HCR RNA-FISH for negative-sense RNA (−N) combined with HCR IF for SND1 in SARS-CoV-2 infected A549ACE2 cells at 8 hpi (MOI = 3 PFU/cell). Representative images are shown. Overlap is quantified by Manders’ co-localization coefficient (n = 6 images). Cells were denatured prior to −N RNA detection; non-denatured cells shown in Figure S3B. Scale bars, 25 μm.