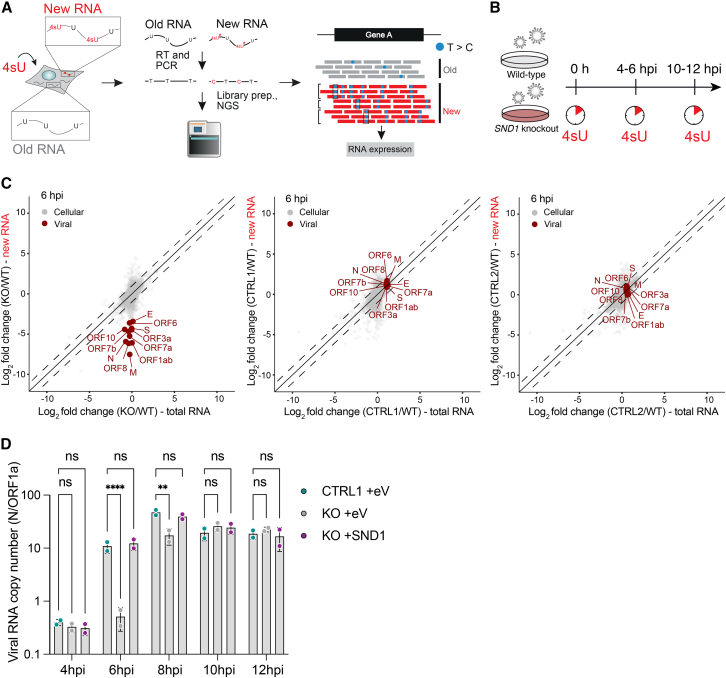
Figure 4.
SND1 is required for nascent SARS-CoV-2 RNA synthesis early during infection
(A) Outline of SLAM-seq method.
(B) Strategy to measure nascent SARS-CoV-2 RNA synthesis in SND1 knockout (KO) and control (CTRL) or wild-type (WT) cells.
(C) SLAM-seq analysis at 6 hpi. Average log2 fold changes in total RNA (x axis) and newly synthesized RNA (y axis) are shown for SND1 KO and CTRL cells, relative to WT cells (n = 2 independent infections).
(D) Absolute quantification of viral RNA copy numbers by digital droplet PCR. Ratio of sgmRNA (N) relative to gRNA (ORF1a) is shown at 4–12 hpi, in SND1 KO and CTRL cells transduced with empty vehicle (+eV) or SND1 (+SND1) as indicated. Cells infected with SARS-CoV-2 at MOI 3 PFU/cell. Values are mean ± SD (n = 2 independent infections). p values determined by two-way ANOVA with Dunnett’s test. ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ns = not significant.
See also Figure S4.