Figure 5.
SND1 interacts with RTC components involved in viral RNA biogenesis
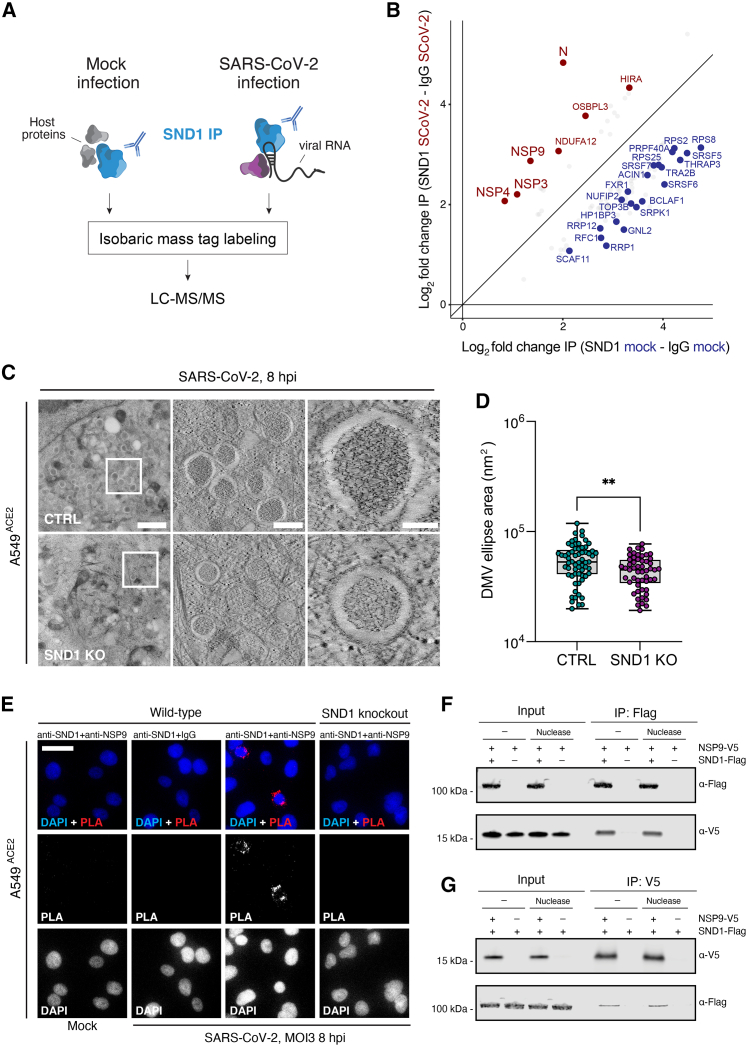
(A) Strategy to globally identify SND1 protein-protein interactome changes upon SARS-CoV-2 infection.
(B) Fold change correlation plot displaying SND1 interacting proteins enriched over IgG control in SARS-CoV-2 infected (SCoV-2, y axis) and uninfected (mock, x axis) A549ACE2 cells (n = 2 independent experiments). Candidates with a fold change > 1.5 or < 0.66 and FDR < 0.2 in infected relative to uninfected cells are displayed. Proteins with a substantial SND1 interaction change (absolute log2 fold change > 1, FDR < 0.05) are highlighted in red and blue.
(C) Computational slices of representative electron tomograms of SND1 knockout (KO) and control (CTRL) cells infected with SARS-CoV-2. Zoom in to region containing DMVs is shown at different magnifications. Left: scale bars, 1 μm; center: scale bars, 250 nm; right: scale bars, 100 nm.
(D) Quantification of cross-sectional DMV area in SND1 KO and CTRL cells. Box: 25th and 75th percentiles. Whiskers: minimum to maximum. Median indicated by line. p value determined by t test, ∗∗p < 0.01.
(E) Proximity ligation assay for SND1 and NSP9. Mock-infected cells are compared to SARS-CoV-2 infected cells. Scale bars, 30 μm.
(F) CoIP western blot analysis for epitope-tagged SND1 and NSP9 proteins expressed in uninfected HEK293T cells with and without nuclease (benzonase) treatment. SND1-FLAG served as bait. Input lysates are shown on the left.
(G) As in (F), but using NSP9-V5 as bait.