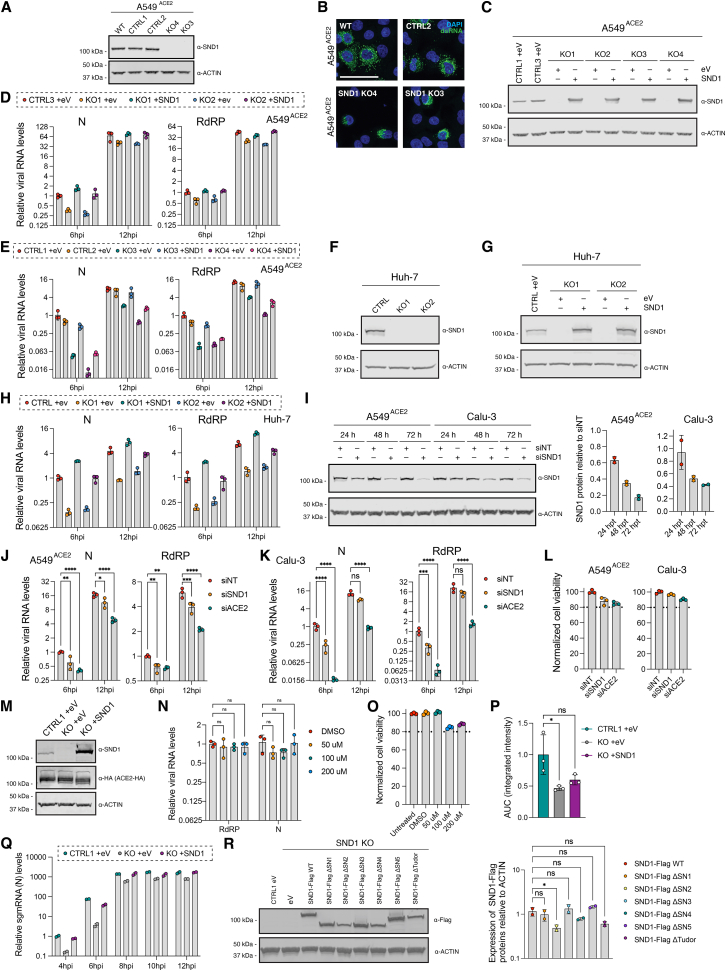
Figure S2.
SND1 is a SARS-CoV-2 host factor, related to Figure 2
(A) Western blot analysis of SND1 knockout (KO), control (CTRL), and wild-type (WT) A549ACE2 cell lines. Expression of SND1 was evaluated relative to actin.
(B) Representative images of IF staining for dsRNA (J2 antibody) using HCR detection in SARS-CoV-2 infected A549ACE2 cells at 8 hpi (MOI = 3 PFU/cell). Scale bars, 50 μm.
(C) Western blot analysis of SND1 KO and CTRL cell lines (A549ACE2) transduced with empty vehicle (+eV), as well as SND1 KO cells transduced with SND1 (+SND1). Expression of SND1 was evaluated relative to actin.
(D) RT-qPCR of SARS-CoV-2 RNA levels (left: N, right: RdRP) at 6 and 12 hpi in SND1 KO and CTRL cells transduced with empty vehicle (+eV) or SND1 (+SND1) as indicated. Two knockout clones shown in Figure 2A were analyzed. A549ACE2 cells were infected at MOI 3 PFU/cell. Quantification relative to 18S rRNA and CTRL at 6 hpi. Values are mean ± SD (n = 3 independent infections).
(E) As in (D), but for two additional knockout clones shown in Figures 2A and S2A.
(F) Western blot analysis of SND1 KO and CTRL cell lines (Huh-7). Expression of SND1 was evaluated relative to actin.
(G) Western blot analysis of SND1 KO and CTRL cell lines (Huh-7), transduced with empty vehicle (+eV), as well as SND1 KO cells transduced with SND1 (+SND1). Expression of SND1 was evaluated relative to actin.
(H) RT-qPCR of SARS-CoV-2 RNA levels (left: N, right: RdRP) at 6 and 12 hpi in SND1 KO and CTRL cells transduced with empty vehicle (+eV) or SND1 (+SND1) as indicated. Huh-7 cells were infected at MOI 3 PFU/cell. Quantification relative to 18S rRNA and CTRL at 6 hpi. Values are mean ± SD (n = 3 independent infections).
(I) Western blot analysis of SND1 siRNA knockdown time course in A549ACE2 and Calu-3 cells at 24, 48, and 72 h post transfection. Expression changes are evaluated relative to non-targeting control and compared to changes in actin. Quantification of SND1 protein levels in two replicate experiments in A549ACE2 and Calu-3 cells is shown on the right. Values are mean ± SD (n = 2).
(J) RT-qPCR of SARS-CoV-2 RNA levels (left: N, right: RdRP) at 6 and 12 hpi in A549ACE2 cells treated with SND1 siRNAs for 72 h. Cells were infected at MOI 3 PFU/cell. Quantification relative to 18S rRNA and CTRL at 6 hpi. Values are mean ± SD (n = 3 independent infections). p values determined by two-way ANOVA with Dunnett’s test.
(K) As in (J), but for Calu-3 cells.
(L) Cell viability assay of SND1 siRNA knockdown experiments at 72 h post transfection. Values are mean ± SD (n = 3).
(M) Western blot analysis of SND1 KO and CTRL cell lines (A549ACE2) transduced with empty vehicle (+eV) or a constitutive lentiviral SND1 expression construct (+SND1). Expression of SND1 and ACE2-HA were evaluated relative to actin.
(N) RT-qPCR of SARS-CoV-2 RNA levels at 6 hpi (left: RdRP, right: N) in A549ACE2 cells treated with DMSO or different concentrations of 2′-deoxythymidine-3′,5′-bisphosphate (pdTp) and infected with SARS-CoV-2 at MOI 3 PFU/cell. Quantification relative to 18S rRNA and DMSO treated cells. Values are mean ± SD (n = 3 independent infections). p values determined by two-way ANOVA with Dunnett’s test.
(O) Cell viability assay of inhibitor treated and untreated cells shown in (N). Values are mean ± SD (n = 3 independent treatments).
(P) Integrated fluorescence intensity-based analysis of three independent time course measurements of SARS-CoV-2-GFP reporter virus growth shown in Figure 2E. Normalization relative to mean of control (CTRL1 +eV). Values are mean ± SD. p values determined by ordinary one-way ANOVA with Dunnett’s test.
(Q) RT-qPCR of N sgmRNA levels at 4–12 hpi in SND1 KO and CTRL cells transduced with empty vehicle (+eV) or SND1 (+SND1) as indicated. A549ACE2 cells were infected with SARS-CoV-2 at MOI 3 PFU/cell. Quantification relative to 18S rRNA and CTRL at 4 hpi. Primers to specifically quantify gRNA and sgmRNA expression levels (Figure 2F) were used. Values are mean ± SD (n = 2 independent infections).
(R) Western blot analysis of SND1 KO and CTRL cells transduced with empty vehicle (eV) or a constitutive lentiviral expression construct encoding the indicated SND1 proteins. Expression of FLAG-tagged SND1 proteins was quantified in duplicates and evaluated relative to actin (right). Values are mean ± SD. p values determined by ordinary one-way ANOVA with Dunnett’s test. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns = not significant.