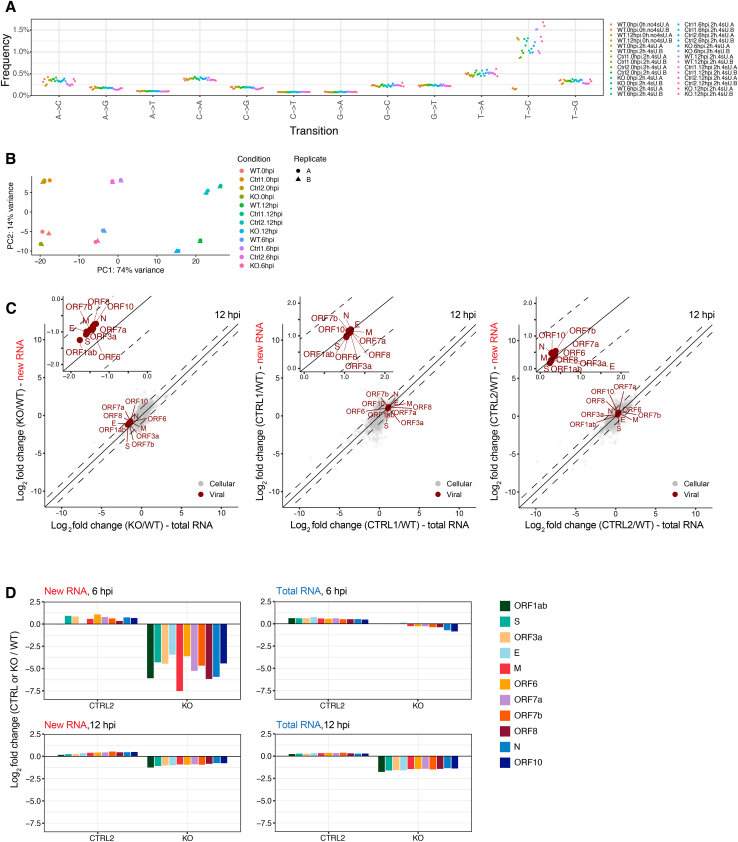
Figure S4.
Measuring changes in viral RNA synthesis with SLAM-seq, related to Figure 4
(A) Mismatch frequencies observed in SLAM-seq libraries.
(B) Principal component analysis of SLAM-seq experiments.
(C) Analysis of SLAM-seq experiments at 12 hpi using GRAND-SLAM. Scatter plot of average log2 fold changes in total RNA (x axis) and newly synthesized RNA (y axis) is shown for SND1 knockout (KO) and control (CTRL) cells, relative to wild-type (WT) cells (n = 2 independent experiments). Insets show a zoom-in view on viral genes for improved visibility.
(D) Bar graph visualization of log2 fold changes for new RNA (left) and total RNA (right) observed in SLAM-seq experiments shown in (C) and Figure 4C for viral genes at 6 (top) and 12 hpi (bottom).