Figure S5.
Expression, localization, and interaction analyses for virus and host proteins, related to Figure 5
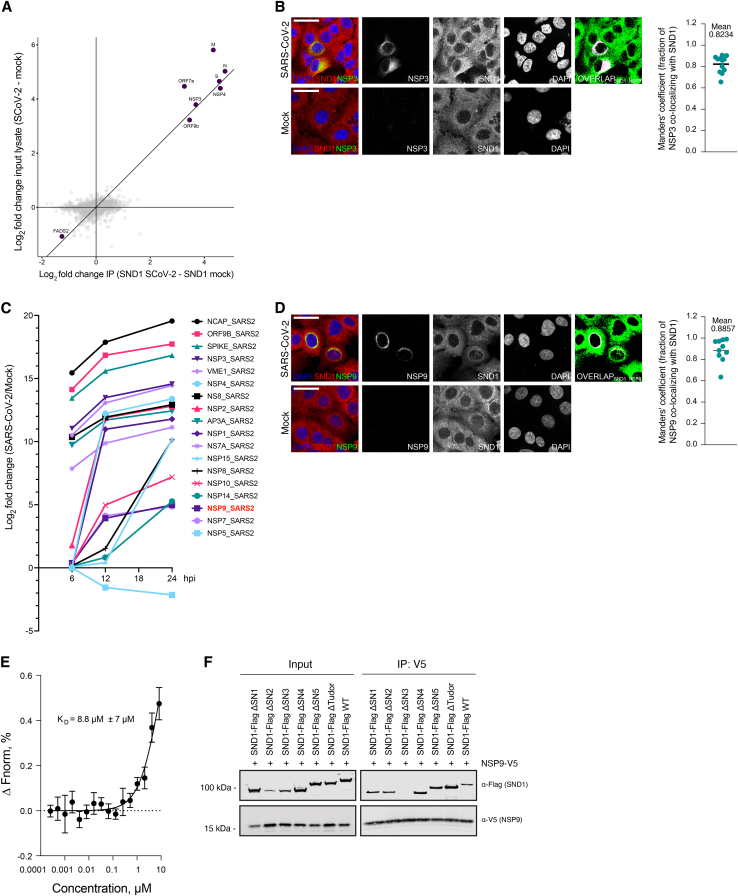
(A) Scatter plot displaying average log2 fold changes in SND1 coIP experiments (x axis) and average log2 fold changes in total proteome measurements of input lysates used for coIP experiments (y axis). SARS-CoV-2 infected cells are compared relative to uninfected cells in coIPs and total proteome measurements (n = 2 independent infections). Proteins that display a significant interaction and expression change (absolute log2 fold change > 1, FDR < 0.05) upon SARS-CoV-2 infection cells are highlighted.
(B) IF staining of SND1 and NSP3 proteins in SARS-CoV-2 infected A549ACE2 cells at 8 hpi (MOI = 10 PFU/cell). Mock-infected cells were stained as controls. Representative images are shown. Overlap between NSP3 and SND1 is quantified by Manders’ co-localization coefficient (right, n = 12 images). Scale bars, 30 μm.
(C) Log2 fold changes for viral proteins observed in total proteome measurements of SARS-CoV-2 infected A549ACE2 cells at different infection time points, as previously reported.18
(D) As in (B), but for SND1 and NSP9. Overlap between NSP9 and SND1 is quantified by Manders’ co-localization coefficient (right, n = 10 images).
(E) Microscale thermophoresis (MST) assay to monitor binding of recombinant NSP9 protein to recombinant SND1 protein in vitro. Unlabeled NSP9 protein (254 pM to 8.33 μM) was titrated against site-specific cysteine-labeled SND1 (10 nM) and thermophoresis was recorded. Change in fluorescence (ΔFnorm) was measured at MST on-time of 10 s. Values are mean ± SD (n = 3 independent measurements).
(F) CoIP western blot analysis for epitope-tagged SND1 and NSP9 proteins exogenously expressed in uninfected HEK293T cells. SND1-FLAG constructs are deleted for indicated protein domains. NSP9-V5 served as bait. Input lysates are shown on the left. Anti-FLAG and anti-V5 antibodies are used to detect tagged SND1 and NSP9 proteins, respectively. SN, SNase.