Figure S6.
Analysis of N and NSP9 RNA binding on viral RNA, related to Figure 6
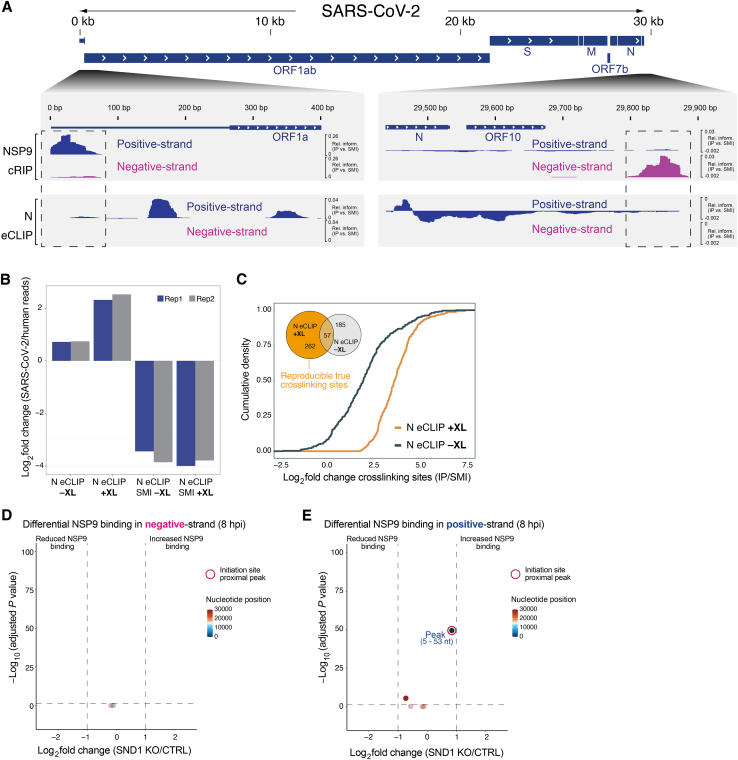
(A) Comparison of the viral RNA binding pattern observed for NSP9 (cRIP, top) and N (eCLIP, bottom) at 12 hpi in A549ACE2 cells. Alignment of strand-separated cRIP and eCLIP data to the SARS-CoV-2 RNA genome is shown. Relative information in IP vs. SMI is calculated at each position (STAR Methods) and displayed for positive-sense RNA (blue) and negative-sense RNA (magenta). Zoom-in views of the 5′ end (left) and the 3′ end (right) of the viral RNA genome are shown and regions of interest are highlighted by dashed box.
(B) Enrichment of reads mapping to the SARS-CoV-2 genome relative to the human genome in N eCLIP experiments with (+XL) and without (−XL) UV-crosslinking, as well as corresponding SMI libraries.
(C) Cumulative density function displaying enrichment of crosslinked nucleotides in N IP relative to SMI experiments with and without UV-crosslinking. Inset shows Venn diagram displaying the overlap of true crosslinking sites reproducibly found in N eCLIP experiments with sites found in N eCLIP experiments without UV-crosslinking.
(D) Volcano plot displaying SND1-dependent changes in NSP9 binding on peak regions relative to non-peak regions in negative-sense RNA at 8 hpi (SND1 KO vs. CTRL). Color scale reflects localization of peak region relative to the 5′ and 3′ end of the viral RNA genome. Peaks with the statistically most pronounced NSP9 binding change are annotated. Initiation site-proximal peaks are encircled.
(E) As in (D), but for positive-sense RNA.