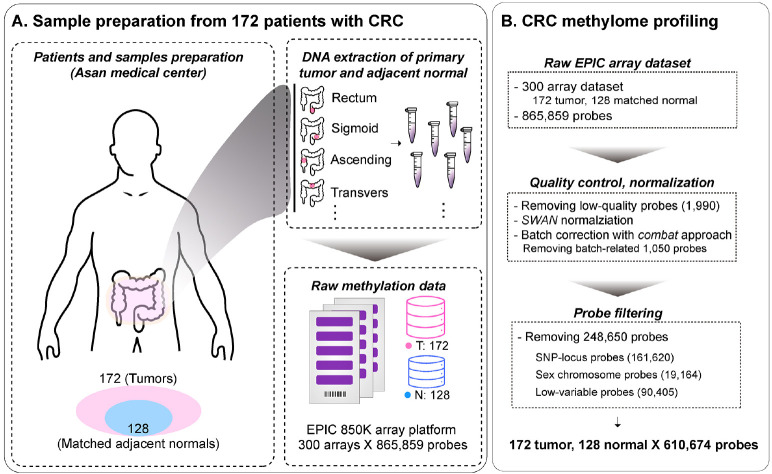
Fig. 1.
Overview of sample preprocessing for CRC methylome profile construction. (A) Preparation of CRC samples for methylome profile. These consisted of 172 tumor samples and 128 adjacent normal samples from patients with CRC at Asan Medical Center. (B) Preprocessing of CRC methylome profile. The minfi pipeline was used for preprocessing, Subset-quantile Within Array Normalization (SWAN) method for normalization, and ComBat approach for batch correction. Poor-performing probes, SNPs, sex chromosomes, and low-variable sites were filtered out. Next, the methylation profiles were finalized. Methylation was quantified at 610,674 probes.