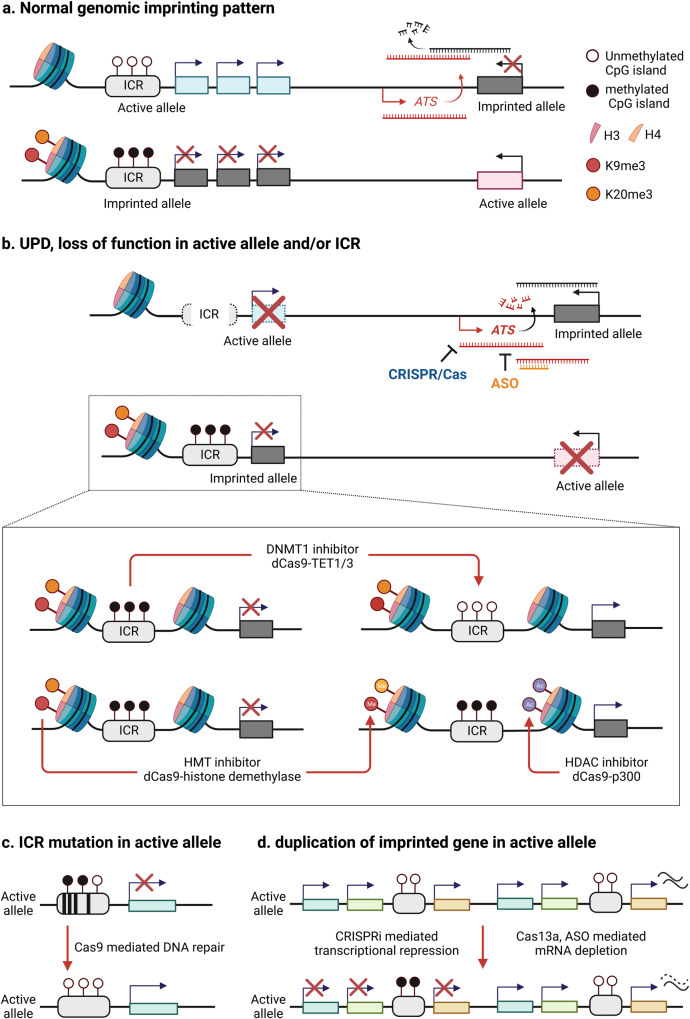
Fig. 1. Epigenetic-based treatment strategies for imprinting disorders.
Schematic diagram describes epigenetic-based therapeutic targets including DNA methyltransferase and histone modifying enzymes for imprinting disorders. a Normal genomic imprinting pattern shows parental origin specific allele silencing. b Uniparental disomy (UPD), deletion or mutation of active allele/imprinting control region (ICR) causes deficiency of normal gene expression. CRISPR/dCas9 or small molecule-mediated reactivation of silenced (imprinted) genes is applicable to recover normal gene expression. c Allele specific CRISPR/Cas9-mediated genome editing can be a tool for correction of ICR mutation resulting in an imprinting defect or epimutation. Designing allele specific gRNA is required to do single nucleotide polymorphism (SNP) analysis to distinguish mat/pat chromosome. d Two copies of imprinted gene in active allele can be repressed by CRISPRi (ex. dCas9-KRAB, DNMT1), CRISPR/Cas13 and ASO mediated mRNA depletion (ASO antisense oligonucleotides, ATS antisense transcripts, DNMT1 DNA methyltransferase 1, HMT histone methyltransferase, HDAC Histone deacetylase). Created with BioRender.com.