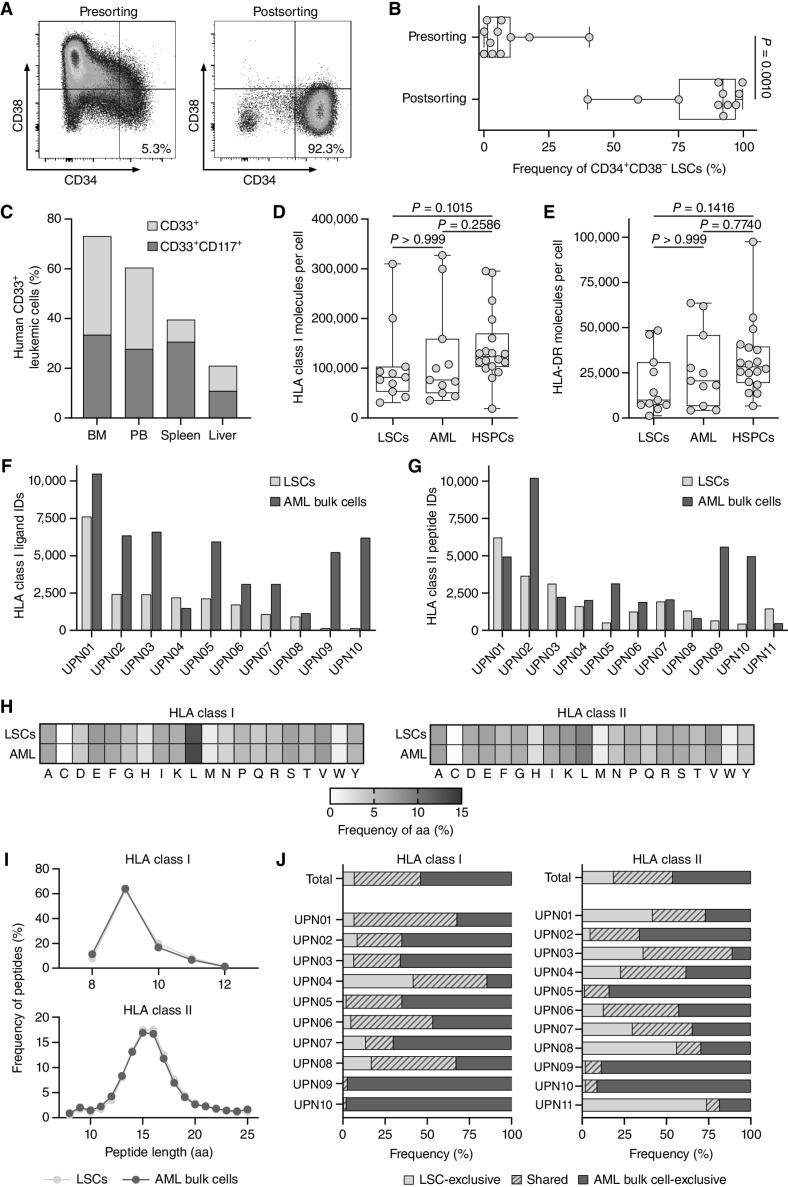
Figure 1.
Immunopeptidome analysis of enriched primary CD34+CD38− LSCs. A, Representative flow cytometry analysis of CD34+CD38− LSC frequencies pre- and postenrichment using magnetic-activated cell sorting (MACS) with the markers CD34 and CD38. B, Frequencies of CD34+CD38− LSCs in primary AML patient samples (n = 11) pre- and postsorting determined by flow cytometry. C,In vivo leukemic engraftment of LSCs (UPN01) in NOD/SCID/IL2Rγnull mice (n = 4). Flow cytometry–based analysis of the frequency of human CD33+ and CD33+CD117+ leukemic cells in the bone marrow, peripheral blood, spleen, and liver of NOD/SCID/IL2Rγnull mice 31 weeks after intrafemoral transplantation of 6 × 105 human CD34+CD38− LSCs. D and E, Surface expression of HLA class I (D) and HLA-DR molecules (E) determined by flow cytometry on AML patient-derived CD34+CD38− LSCs (n = 11) and CD34+CD38+ AML cells (n = 11) and on HV-derived CD34+ HSPCs (n = 18, thereof n = 7 hematopoietic stem cell apheresis from G-CSF mobilized blood donations of patients with nonhematologic malignancies). F and G, Number of mass spectrometric identified HLA class I- (F) and HLA class II-presented peptides (G) on LSCs and corresponding AML bulk cells (n = 10 for HLA class I, n = 11 for HLA class II). H, Amino acid distribution within the LSC- and AML bulk cell-derived HLA class I (left) and HLA class II (right) immunopeptidomes based on the unique peptide identifications in each cohort. I, Peptide length distribution of HLA class I ligands (top) and HLA class II peptides (bottom) in the immunopeptidome of LSCs and corresponding AML bulk samples. J, Overlap analysis of HLA class I ligand (left) and HLA class II peptide (right) identifications of LSCs and corresponding AML bulk samples on patient-individual and cohort-wide level. In B, D, and E, data points represent individual samples. Boxes represent median and 25th to 75th percentiles, whiskers are minimum to maximum. C, Data are presented as bar graphs with mean. B, Paired Wilcoxon signed rank test. D and E, Kruskal–Wallis test. Abbreviations: BM, bone marrow; PB, peripheral blood; ID, identification; UPN, uniform patient number; aa, amino acid.