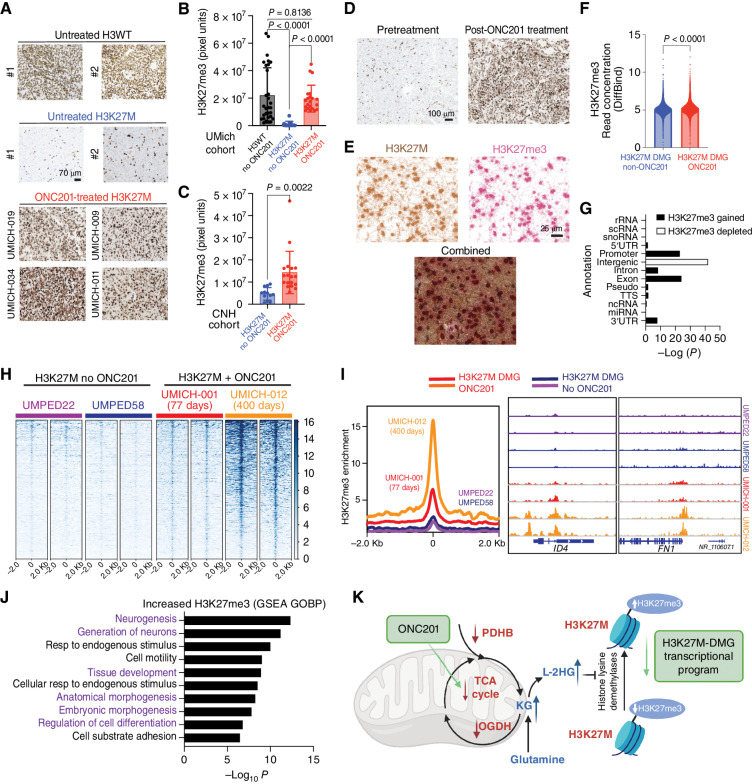
Figure 7.
ONC201 treatment increases genomic H3K27me3 in H3K27M-DMG patient tumors. A, Representative images of tumor samples stained for H3K27me3 from untreated H3WT, untreated H3K27M-DMG, and ONC201-treated H3K27M-DMG patient tumor samples. B, Quantification of H3K27me3 (3 regions were captured in multiple tumor areas in a blinded manner/case) in H3WT non–ONC201-treated (n = 10, black), H3K27M non–ONC201-treated (n = 11, blue), or H3K27M ONC201-treated (n = 6, red) patient samples from the UMich Cohort. Data were analyzed using ANOVA. C, Quantification of H3K27me3 (3 regions were captured in multiple tumor areas in a blinded manner/case) in H3K27M non–ONC201-treated (n = 4, blue) or H3K27M ONC201-treated (n = 6, red) independent, nonoverlapping patient samples from the Children's National Hospital (CNH) Cohort. Data were analyzed using an unpaired, two-tailed, two-sided, nonparametric Mann–Whitney test. D, Representative images of H3K27M-DMG tumor samples from the UMich cohort stained for H3K27me3 from the same patient pretreatment (biopsy) or post-ONC201 treatment (autopsy). Data were analyzed using unpaired, two-tailed, two-sided, Student t test. E, Representative images from an ONC201-treated H3K27M-DMG tumor sample from the UMich Cohort stained with combined IHC for H3K27M (brown) and H3K27me3 (red). Images show brown or red chromogens or an overlay of both. F, Overall genome-wide H3K27me3 in ChIP-seq from H3K27M-DMG tumor samples treated with or without ONC201 treatment (n = 2 patients per condition). Data were analyzed using an unpaired, two-tailed, two-sided, nonparametric Mann–Whitney test. G, Comparison of genomic H3K27me3 in H3K27M-DMGs tumor samples derived from patients treated with (n = 2) or without (n = 2) ONC201 from F. H, Heat maps showing genomic H3K27me3 levels (± 2 Kb from peak center) in H3K27M-DMG tumor samples with (patients UMICH-001 and UMICH-012) or without (patients UMPED22 and UMPED58) ONC201 treatment. Each set of two heat maps represents replicates obtained from different tumor regions for each patient. I, Left, overall peak representation of genomic H3K27me3 in ONC201-treated (UMICH-001, orange; and UMICH-012, red) and non–ONC201-treated H3K27M-DMGs (UMPED22, purple; and UMPED58, blue). Right, representative H3K27me3 tracks for genes ID4 and FN1 from each tumor sample. J, GSEA of genes with significantly increased H3K27me3 in ONC201-treated versus untreated patients from F–I. K, Schema of the overall proposed mechanism of ONC201 impact on metabolic and epigenetic signaling in H3K27M-DMGs.