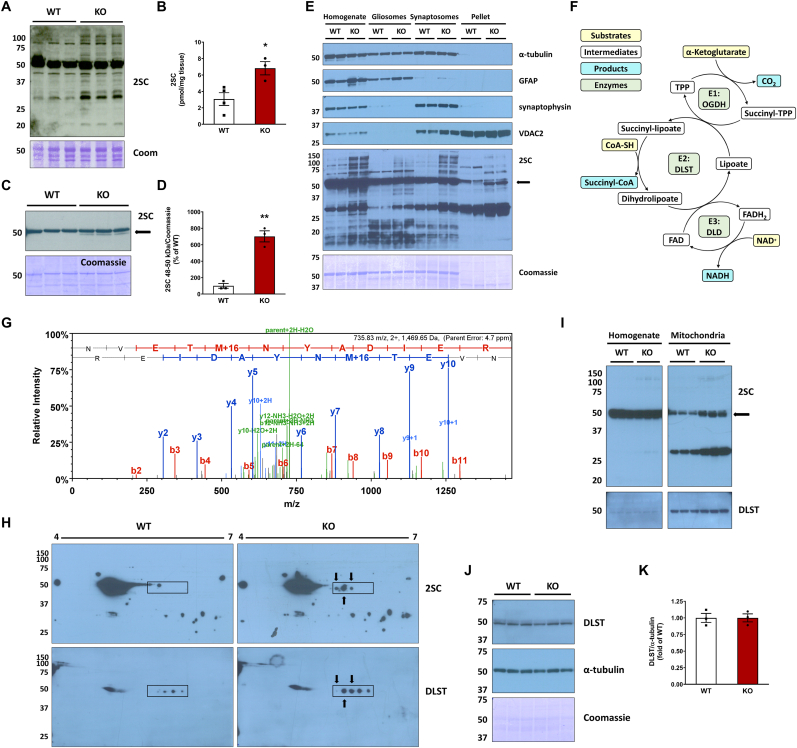
Fig. 1.
Identification of dihydrolipoyllysine-residue succinyltransferase (DLST) succination in the brainstem (BS) of the Ndufs4 KO mouse.A, Total levels of protein succination (2SC) in brainstem (BS) homogenates from WT and Ndufs4 KO mice, note the abundance of succinated tubulin in WT and KO at ∼50 kDa. Ndufs4 KO BS show enrichment of several additional succinated proteins vs. WT mice (representative blot, n = 3/group). B, LC-MS/MS quantification of total 2SC levels in WT and Ndufs4 KO brainstem (n = 3–4/group). C, Detection and D, densitometric quantification of a distinct succinated band at ∼48–50 kDa (arrow) in Ndufs4 KO BS following cytosolic tubulin depletion (n = 3). E, Characterization of the purity of total tissue protein homogenates, gliosomes, synaptosomes, and a mitochondria-rich pellet fraction obtained from BS by analysis of α-tubulin, GFAP, synaptophysin and VDAC2 expression. In the 2SC panel, a ∼48–50 kDa succinated band is present in the pellet fractions (2SC panel, KO lanes, arrow). F, Structure and function of the α-ketoglutarate dehydrogenase complex (KGDHC). For each subunit the substrates are highlighted in yellow, intermediates/cofactors in white and products in light blue and the enzymatic components of the complex in light green. G, MS/MS spectrum of the peptide NVETMoxNYADIER corresponding to DLST in a ∼48–50 kDa band isolated from the mitochondria-rich pellets used in C. H, Co-localization (arrows) of succinated spots with DLST-immunoreactive spots in the region of ∼48–50 kDa and pH 5.5–6.3 (rectangular area) in Ndufs4 KO BS. I, Analysis of BS tissue homogenates, where tubulin precludes DLST detection, alongside purified mitochondrial fractions showing a succinated band at ∼48–50 kDa in KO lanes (2SC panel, arrow), which overlapped with DLST detection. J, Total DLST and α-tubulin content in BS homogenates of WT and Ndufs4 KO mice. K, Quantification of the DLST band intensity in J (n = 3/group). See also Fig. S1.In A, C, and E, H, I, J molecular weight markers are shown on the left side. Coomassie staining was used to verify even load of the lanes. In B, D, K, results expressed as mean ± SEM were compared by unpaired t-test (*P < 0.05, **P < 0.01). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)