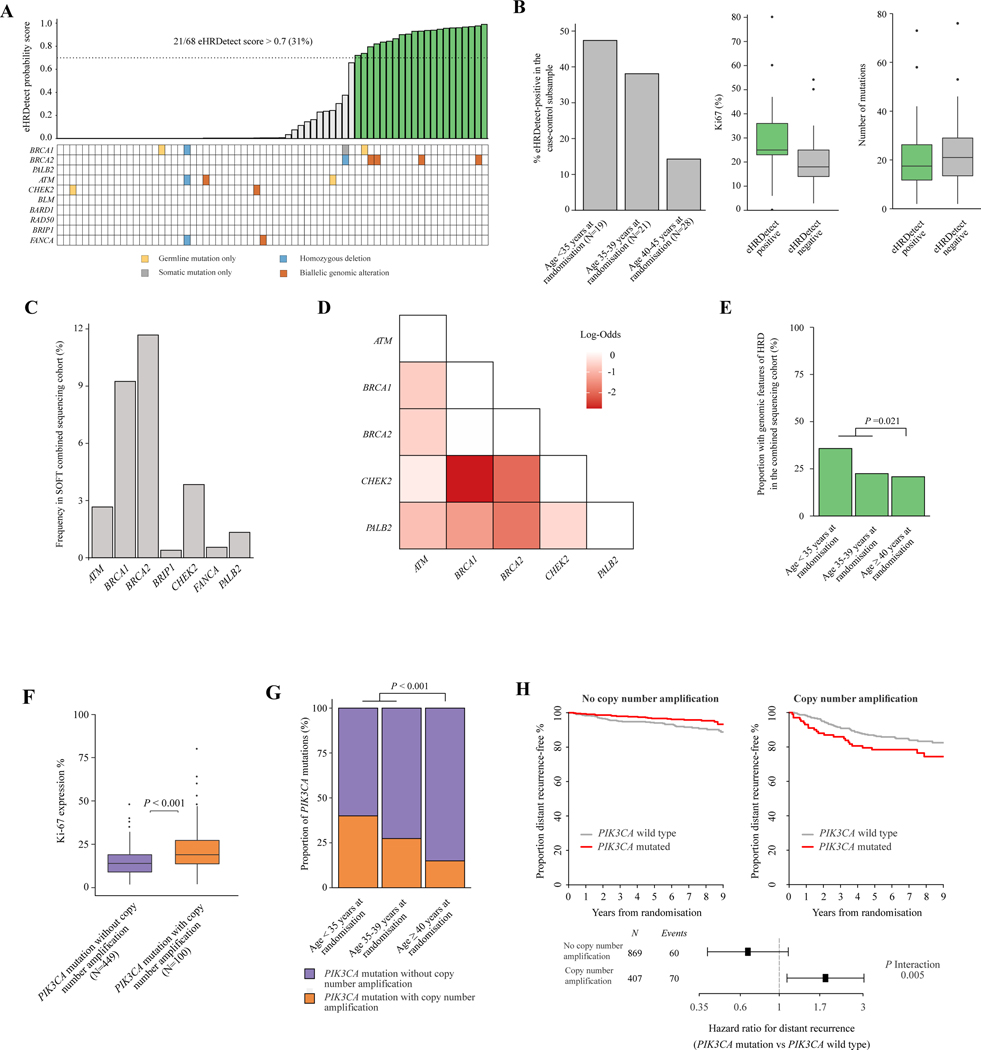
Figure 4. Genomic features of HRD and high-risk PIK3CA mutations in young patients.
(A) Plot demonstrating eHRDetect probability score (top panel) in 68 assessable patients in the case-control subsample who underwent paired tumour-normal whole-exome sequencing. Green bars indicate eHRDetect score above the 0.7 probability score threshold, termed eHRDetect positive. Genomic alterations in HRD-related genes (bottom panel) are also shown. (B) Plots demonstrating the associations between eHRDetect-positive score with age at randomisation, Ki-67 expression level, and number of somatic mutations in the case-control subsample who were assessable for eHRDetect (n = 68). (C) Bar plot demonstrating the frequency of patients with genomic features of HRD according to age at randomisation in the SOFT combined sequencing cohort (n = 1276). Genomic features of HRD included eHRDetect positivity and/or genetic alterations in HRD-related genes. (D) Pairwise analysis of HRD-related genes in the SOFT combined sequencing cohort (n = 1276). Only genes with ≥10 patients with genetic alterations were included. Fisher’s exact test was applied to each genetic alteration pair. Multiple testing correction using false discovery rate was applied. Only log-odds with a false discovery rate of <0.2 are displayed. Burgundy colour indicates an association with mutual exclusivity. (E) Bar plot demonstrating the proportion of tumours with genomic features of HRD according to age at randomisation in the SOFT combined sequencing cohort (n = 1276). Genomic features of HRD included eHRDetect positivity and/ or genetic alterations in HRD-related genes. (F) Boxplot demonstrating the association between concurrent copy number amplification status with PIK3CA mutations and Ki-67 expression levels in the SOFT combined sequencing cohort (n = 1276). (G) Bar plot demonstrating the frequency of patients with a PIK3CA mutation and concurrent copy number amplification according to age at randomisation in the SOFT combined sequencing cohort (n = 1276). (H) Kaplan–Meier plots and forest plots estimating the rate of freedom from distant recurrence according to PIK3CA mutation status and copy number amplification status in the SOFT combined sequencing cohort (n = 1276).
HRD, homologous recombination deficiency; SOFT, Suppression of Ovarian Function Trial.