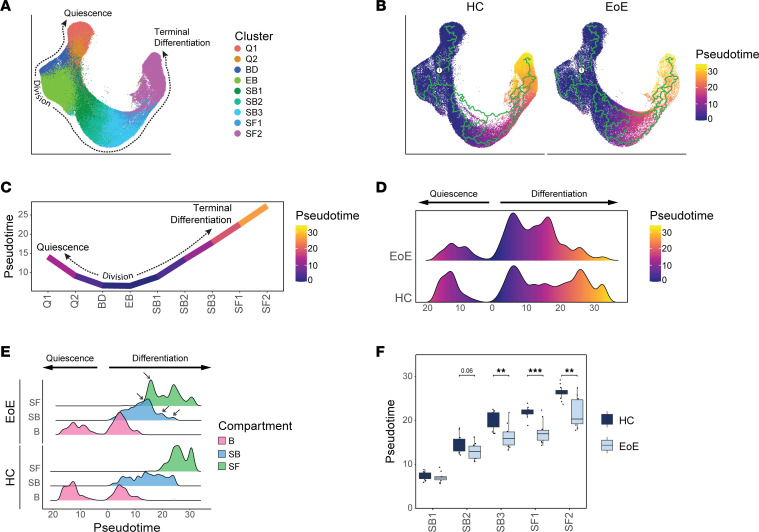
Figure 7. Pseudotemporal trajectory analysis of EEC reveals a shift in global differentiation toward basal identity in EoE.
(A) UMAP showing merged scRNA-Seq data sets of EEC from HC and EoE, colored by integrated clusters. (B) Pseudotime trajectory analysis calculated using HC and EoE samples, with cycling cells as the pseudotime origin. (C) Average pseudotime values across epithelial clusters. Pseudotime is rooted in cycling cells and progresses toward either quiescence or terminal differentiation. (D) Ridgeline plot illustrating the pseudotime value distribution between EEC in HC and EoE. (E) Ridgeline plots depicting pseudotime value distribution between epithelial compartments in HC and EoE. Arrows indicate peaks with differential density between EoE and HC. (F) Box plot showing the distribution of pseudotime values for every suprabasal and superficial cell cluster in HC or EoE. Boxes indicate quartiles, whiskers indicate minima/maxima separated by 1.5 times the interquartile range, and lines through each box indicate median pseudotime value. Indicated P values were determined using Wilcoxon signed-ranked test with Benjamini & Hochberg adjustment for multiple comparisons. **P ≤ 0.01, ***P ≤ 0.001. B, Basal; Q, Quiescent; BD, Basal_Dividing; EB, Epibasal; SB, Suprabasal; SF, Superficial.