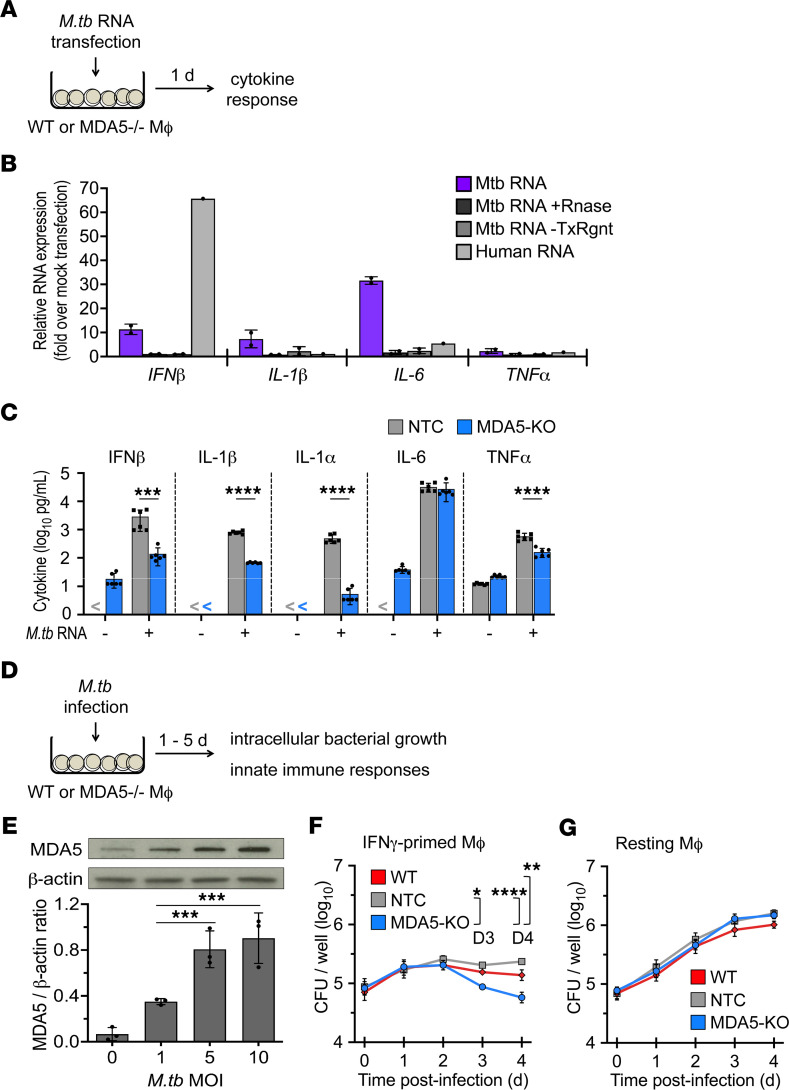
Figure 1. Cytosolic RNA sensor MDA5 modulates host cytokine response to M. tuberculosis–derived RNA and promotes M. tuberculosis intracellular growth in macrophages.
(A) Schematic diagram of experimental design to identify host factors involved in cytosolic surveillance of M. tuberculosis RNA in macrophages. (B) Resting primary human monocyte-derived macrophages were transfected for 24 hours with total M. tuberculosis RNA, total M. tuberculosis RNA treated with RNaseV (+RNase), mock transfection of total M. tuberculosis RNA (–TxRngt), or total human RNA. IFN-β, IL-1β, IL-6, and TNF-α RNA levels were assessed by reverse transcriptase qPCR. RNA levels were normalized to PolR2A. Data are presented as fold-change relative to no-transfection control (mean ± SD, n = 2 biological replicates, 2 independent experiments). (C) CRISPR/Cas9–mediated Mda5-knockout (Mda5-KO) or CRISPR/Cas9 nontarget control (NTC) J774.1 cells were IFN-γ–primed and transfected with total M. tuberculosis RNA for 24 hours. IFN-β, IL-1β, IL-6, and TNF-α levels in the culture medium were quantified by multiplex immunoassay (Luminex) (mean ± SD, n = 6 biological replicates from 3 independent experiments; “<” indicates below limit of quantification). ***P < 0.001, ****P < 0.0001 by 2-way ANOVA with Tukey’s posttest. (D) Schematic diagram of experimental design to evaluate the role of Mda5 in innate immune responses to M. tuberculosis infection. (E) MDA5 and β-actin protein levels in WT J774.1 cells at 24 hours after infection with M. tuberculosis at the indicated MOI were detected by immunoblot analysis and quantified by densitometry (mean ± SD, n = 3 biological replicates, 3 independent experiments). ***P < 0.001 by 1-way ANOVA with Tukey’s posttest. (F and G) Growth kinetics of M. tuberculosis (MOI of 1:5) in IFN-γ–primed (F) and resting (G) WT, NTC, and Mda5-KO J774.1 cells. Data are mean CFU ± SEM for each time point (n = 4 biological replicates, 2 independent experiments). *P < 0.05, **P < 0.01, ****P < 0.0001 by 2-way ANOVA with Tukey’s posttest.