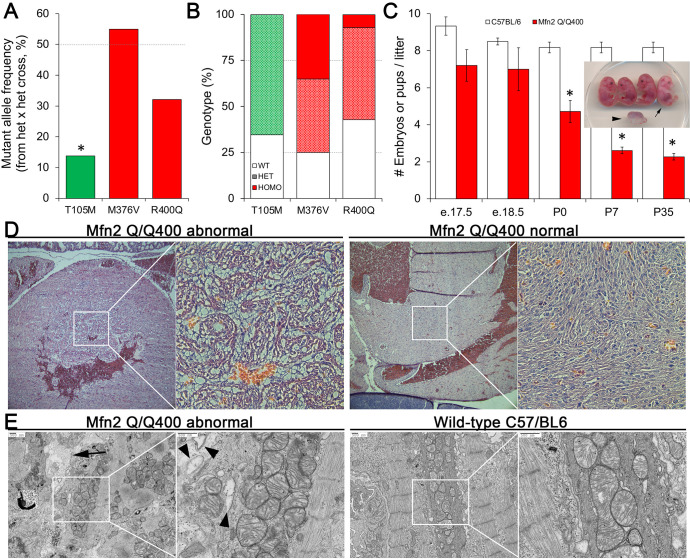
Figure 4. Perinatal cardiomyopathy in Mfn2 Q/Q400 mice.
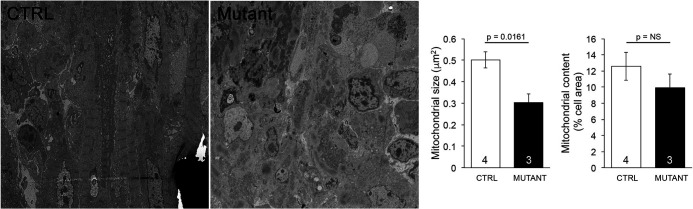
(A, B) Mutant allele frequencies (A) and genotypes (B) from heterozygous crosses of Mfn2 T105M, M376V, and R400Q knock-in mice (CRISPR/Cas9 knock-in strategies are given in the figure supplement). Expected values are shown as dotted lines. Homozygous mice were absent for T105M and fewer than expected for R400Q. (C) Fetal and early postnatal lethality of homozygous R400Q knock-in mice (Q/Q400) reported as mice/litter at different time points. Controls (white) are C57/Bl6. Inset shows an E18.5 Q/Q400 litter having a degenerated fetus (arrowhead) and a live, non-viable fetus (arrow). (D) Hematoxylin and eosin-stained sections of left ventricular myocardium from P0 Q/Q400 mice with (left) and without (right) cardiomyocyte vacuolization. (E) Ultrastructural studies of Q/Q400 myocardium revealing myofibrillar degeneration (arrow), empty ‘ghost’ mitochondria (arrowheads) and mitochondrial fragmentation; wild-type control is shown for comparison on the right. *p<0.05 vs. expected (A, Fisher’s exact test) or control (C, Student’s t-test).