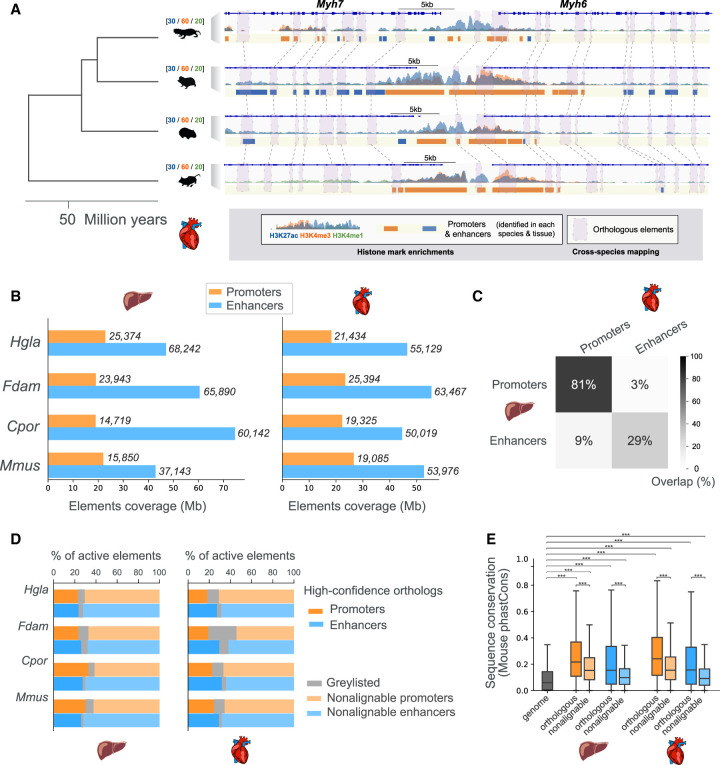
Figure 1.
Comparative epigenomics of mole-rat promoters and enhancers in the heart and liver. (A) Epigenomic profiling and cross-mapping approach exemplified by the Myh6/Myh7 heart locus. H3K27ac (blue), H3K4me3 (orange), and H3K4me1 (green) histone mark enrichment in the heart are shown for each of the four species: (Hgla) naked-mole rat, (Fdam) Damaraland mole-rat, (Cpor) guinea pig, and (Mmus) mouse. Scales above species names indicate fold enrichment over input (averaged across replicates). Note that we use the same scale across species to facilitate direct visual comparison of histone mark enrichments. Identified promoters and enhancers are represented by orange and blue boxes, respectively. Purple boxes connected by dashed lines correspond to orthologous promoters and enhancers in each species. (B) Coverage and numbers of promoters and enhancers by species and tissue. Bars correspond to total genomic coverage; the number of elements are indicated at the right of bars. (C) Overlap between promoters and enhancers across the liver and heart tissues, averaged over the four species. Percentages represent the fraction of overlapping elements in each category. (D) Percentage of promoter and enhancer elements containing high-confidence orthologous regions across the four species (dark orange and blue bars). Lighter shade bars indicate promoters and enhancers with no high-confidence orthologs (nonalignable). Graylisted regulatory elements correspond to elements with elevated read density in input ChIP (Methods). (E). Sequence conservation of orthologous and nonalignable promoters and enhancers in the mouse. Orthologous mouse elements have significantly higher phastCons sequence conservation scores than those of nonalignable elements. Both orthologous and nonalignable mouse elements have sequences significantly more conserved than the genomic background: Mann–Whitney U tests, with Benjamini–Hochberg correction for multiple testing, (***) corrected P-values < 0.001. See also Supplemental Figures S1 and S2 and Supplemental Table S1.