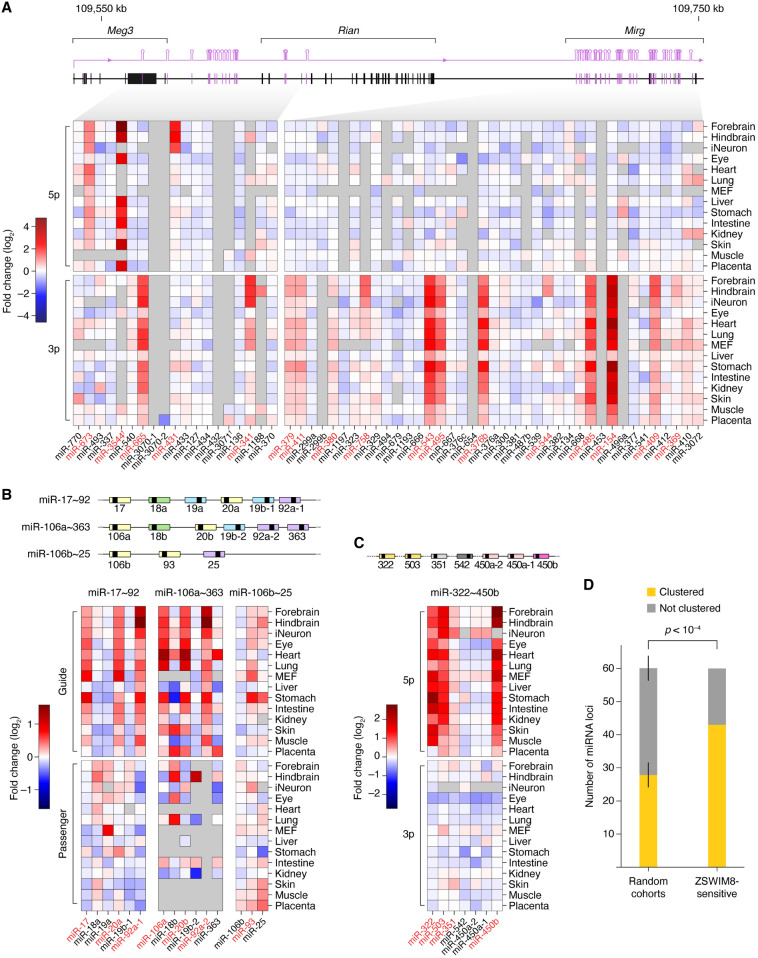
Figure 6.
Genomic organization of ZSWIM8-sensitive miRNAs. (A) Clustered miRNA genes of the imprinted Dlk1–Dio3 locus, which produces a preponderance of ZSWIM8-sensitive miRNAs. Shown are fold-changes in miRNA levels observed in Zswim8−/− tissues, relative to Zswim8+/− tissues from E18.5 embryos, as well as in MEFs and iNeurons (Shi et al. 2020). The miRNAs are ordered by position within the cluster and organized by strand. Each plotted value is the average of two biological replicates. Red text denotes ZSWIM8-sensitive miRNAs; dagger, miRNA apparently derived from the antisense strand of the paternal allele. (B) The miR-17∼92 cluster and two paralogous clusters. Otherwise, as in panel A. (C) The miR-322∼450b cluster. Otherwise, as in panel A. (D) A tendency of ZSWIM8-sensitive miRNAs to derive from clustered miRNA genes. The bar plots show the proportion of miRNA loci produced from a miRNA cluster for the set of 61 loci encoding ZSWIM8-sensitive miRNAs compared with random cohorts of the same size drawn without replacement from the set of all miRNA loci. Error bars, SD across 10,000 random cohorts. The P-value was calculated by hypergeometric test.