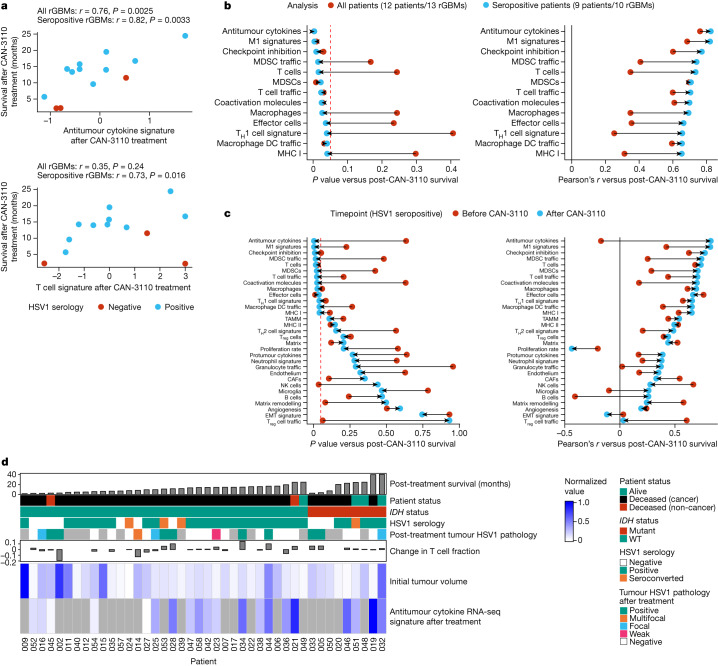
Fig. 5. Survival correlation between immune transcript signature programs in HSV1-seronegative and HSV1-seropositive patients.
A total of 13 paired IDHWTrGBMs with good-quality RNA was analysed by bulk RNA transcriptomics. Transcriptomic signatures for different biological programs were estimated for each sample, and these signatures were assessed for correlation with survival after CAN-3110 treatment either in all patients or only in patients who were HSV1 seropositive before or after CAN-3110 treatment. a, Example of two immune signatures (antitumour cytokine and T cell signatures) that are strongly correlated with survival after CAN-3110 treatment when analysed in HSV1 seropositive patients. Pearson’s r correlation coefficients and P values (two-sided, based on t-distribution) are shown above the plots. Importantly, these signatures did not appear to be significantly confounded by the tissue collection timepoint (Extended Data Fig. 12c). b, The change in Pearson’s correlation P (left) (two-sided, based on t-distribution) and r (right) values when correlations between post-treatment immune signatures and survival were performed in all patients (red points) or in only HSV1-seropositive patients (teal points). Only gene signatures that reached P ≤ 0.05 (dashed red line) in either analysis were plotted. DCs, dendritic cells; MDSCs, myeloid-derived suppressor cells; TH1, T helper 1. c, The change in Pearson’s correlation P (left) (two-sided, based on t-distribution) and r (right) values for pre-treatment (red points) and post-treatment (teal points) samples from HSV1-seropositive patients. This panel includes all of the analysed RNA-seq gene signatures. The dashed red line indicates P = 0.05. CAFs, cancer-associated fibroblasts; EMT, epithelial–mesenchymal transition; NK cells, natural killer cells; TAMM, tumour-associated monocyte/macrophage; Treg cells, regulatory T cells. d, Combined data for all of the patients in the study, including survival after CAN-3110 treatment, HSV1 serology, HSV1 tumour pathology, T cell fraction changes based on TCRβ DNA-seq, initial tumour volumes and bulk RNA-seq-based antitumour cytokine signature scores. The grey boxes indicate missing data. For b and c, HSV1 serology remained unchanged after CAN-3110 treatment for all of the patients, and one patient (045) was omitted from the analysis due to early non-GBM mortality.