Fig. 1.
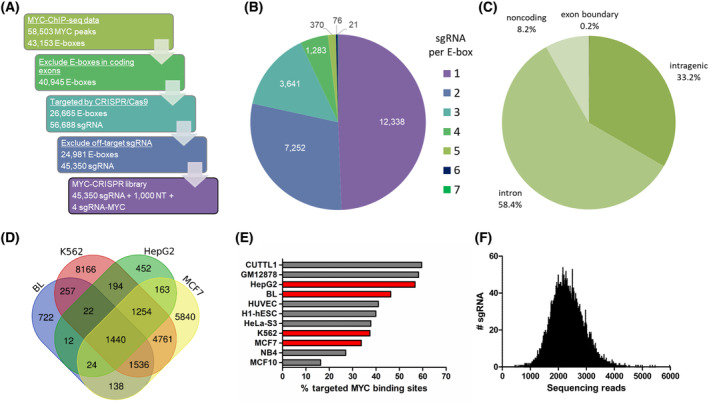
Design and generation of the MYC‐EBOX‐CRISPR library for genome‐wide disruption of MYC binding sites. (A) Library was designed based on publicly available MYC‐ChIP‐seq data in MYC‐dependent K562, MCF7, HepG2, and Burkitt lymphoma (BL) cell lines. After excluding E‐boxes in coding exons, all possible sgRNAs targeting remaining E‐boxes were designed, based on the presence of PAM sequence. sgRNAs with predicted off‐target binding were filtered out. The final library contains 43 350 sgRNAs targeting E‐boxes, 1000 non‐targeting (NT) sgRNAs as a negative control, and four sgRNAs targeting MYC as a positive control. (B) Number of sgRNA constructs per E‐box. (C) Genomic location of E‐boxes targeted by the library. (D) Overlap of targeted E‐boxes for selected cancer cell lines. (E) Percentage of MYC binding sites targeted by the MYC‐EBOX‐CRISPR library in various cell lines, based on available MYC‐ChIP‐Seq data. In red, are cell lines for which the library was designed. (F) Distribution of sgRNA constructs in the MYC‐EBOX‐CRISPR plasmid library determined by NGS. All sgRNAs were present in the library.
