Fig. 6.
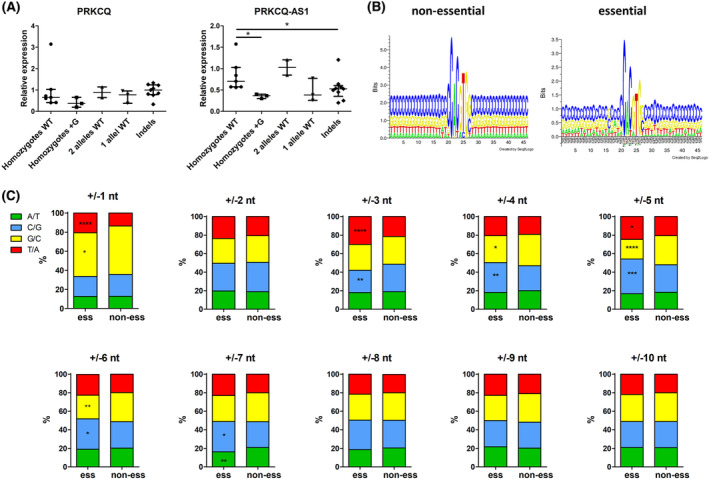
Grammar of the E‐box sequence context. (A) Expression of two genes adjacent to an E‐box on chromosome 10 (chr10_BS212) was examined in monoclonal cell lines derived from K562 cells transduced with an sgRNA targeting this E‐box. A spectrum of clones with various modifications of the E‐box was obtained, including wild‐type (WT) homozygotes (n = 7; K562 cells are triploid); homozygotes with the +G insertion after E‐box (n = 3); clones with two WT alleles and one mutated allele (n = 2); clones with one WT allele and two mutated alleles (n = 3); and clones with various indels on all three alleles (n = 9). Median with interquartile range is shown; *, P < 0.05, Kruskal–Wallis test with Dunn's post‐test. (B) Sequence logo (created using Seq2Logo) of the E‐boxes and 20 nt flanking sequences for non‐essential E‐boxes (left, n = 24 705) and E‐boxes essential in at least one cell line (right, n = 276). (C) Frequency of up to 10 nt upstream/downstream flanking essential (n = 276) and non‐essential (n = 24 705) E‐boxes. Since E‐boxes are (quasi)palindromic and can be read on either strand, G at +1 equals C at −1, etc. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; chi‐squared goodness‐of‐fit test.
