Figure 5. CRISPR‐Cas‐based genome editing tools.
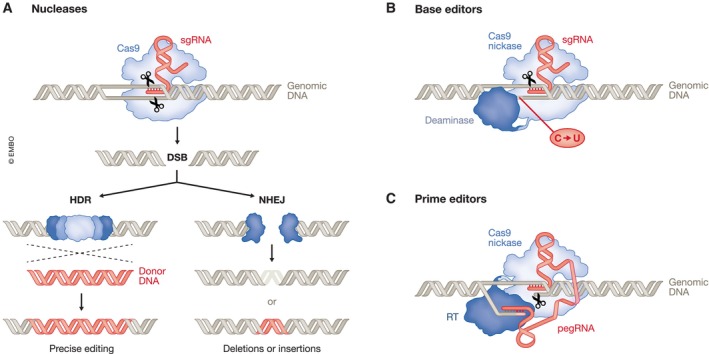
(A) Cas9 nucleases are guided to their target region by a small guide RNA (sgRNA). There they generate double‐strand breaks (DSBs) that are repaired by either homology‐directed repair (HDR) or non‐homologous end joining (NHEJ). HDR repairs DSBs by homologous recombination using a donor DNA template. This can be the sister chromatid when it is present during the late S or G2 phase of the cell cycle or an exogenously supplied donor that supports gene correction. In contrast, NHEJ, the dominant path for DSB repair, often leads to uncontrolled nucleotide deletions or insertions. (B) Base editors mediate targeted single‐nucleotide conversions using a fusion between a Cas9 nickase and a deaminase domain, which modifies single bases through deamination. Nicking of the nondeaminated strand biases cellular DNA repair to replace the unedited strand, thereby resolving the mismatch and leading to stable conversion. Base editors are able to mediate targeted C > U, A > G, C > G, A > I and C > T conversions. (C) Prime editors consists of a reverse transcriptase (RT) fused to a Cas9 nickase. The Cas9 nickase binds to and nicks the non‐target DNA strand and the reverse transcriptase subsequently uses the resulting free 3′ end to copy the sequence of the prime‐editing guide RNA (pegRNA).
