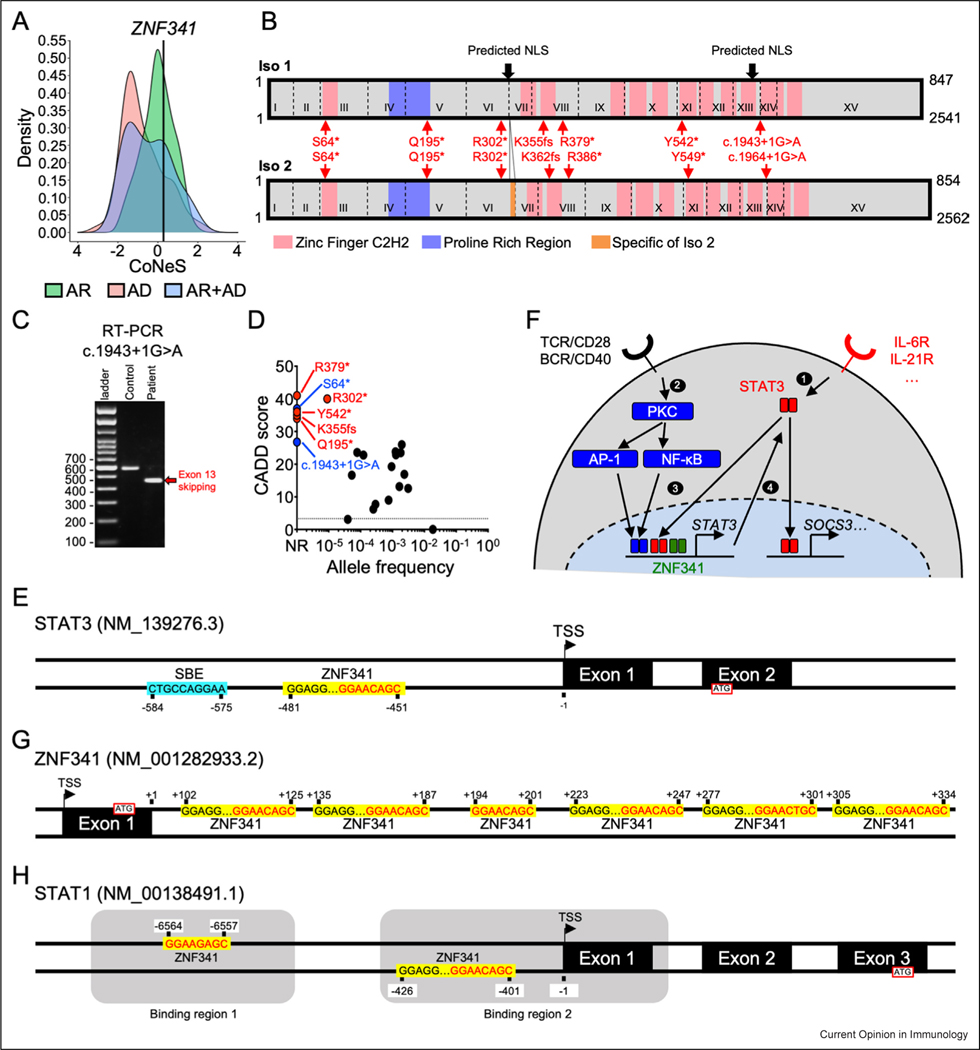
Figure 1.
ZNF341 isoforms, population genetics, schematic signaling model, and DNA binding sites in STAT1, STAT3 and ZNF341 genes. (a) The CoNeS score of ZNF341 is compatible with an AR trait. (b) Schematic representation of ZNF341 isoforms 1 (top) and 2 (bottom). The exons are indicated by Roman numerals, and the exon boundaries are indicated with dashed lines. Predicted nuclear localization sequences are indicated by black arrows. The seven reported mutations underlying HIES are indicated in red for both isoforms. (c) Reverse transcription PCR (Polymerase Chain reaction) showing that the newly reported c.1943+1G > A mutation is responsible for exon-13 skipping. (d) Population genetics of ZNF341. All HIES variants are highly deleterious and only the R302* mutant has been reported in the heterozygous state in a public database. CADD: combined annotation-dependent depletion score. (e) Schematic representation of the STAT3 promoter. The SBE (STAT3 binding element) is indicated by a blue box. ZNF341-binding sites are indicated by yellow boxes. The TSS (Transcription start site) is indicated. The position of each binding site is indicated relative to the TSS. (f) Model for the ZNF341-dependent signaling pathway. BCR: B-cell receptor. (g–h) Schematic representation of the ZNF341 exon 1 and intron 1 (g), and STAT1 promoter (h), as in E. The STAT1 promoter contains two binding regions indicated by gray boxes.