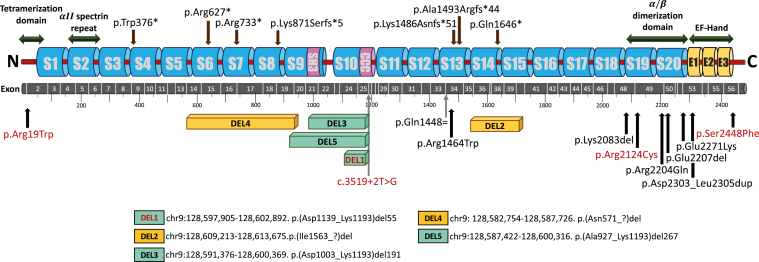
Figure 2.
Schematic structure of SPTAN1 gene and its coding protein highlighting variants identified in this study. Coding exon numbers (NM_001130438.3) are reported on the gray bar. Truncating variants are indicated on the top. Missenses, in-frame deletion/insertion, and splice variants are on the bottom. Deletion 1, 3, and 5 (green) remain in frame, whereas predictions for deletions 2 and 4 (orange) are not available. p.Gln1448= (c.4344G>A) is predicted to affect exon 33 donor splice site, based on maxENTScan (predicting splice sites using ‘Maximum Entropy Principle’) (maxENT score wild-type 6.99 → 3.84 mutant). The splice altering variant (NC_000009.12(SPTAN1_v001):c.3519+2T>G) predicted to alter exon 25 canonical donor splice site (maxENT score wild-type 10.28 → 2.63 mutant). Variants identified in patients presenting with HSP/HA are highlighted in red. All other variants are represented in black.