Figure 1.
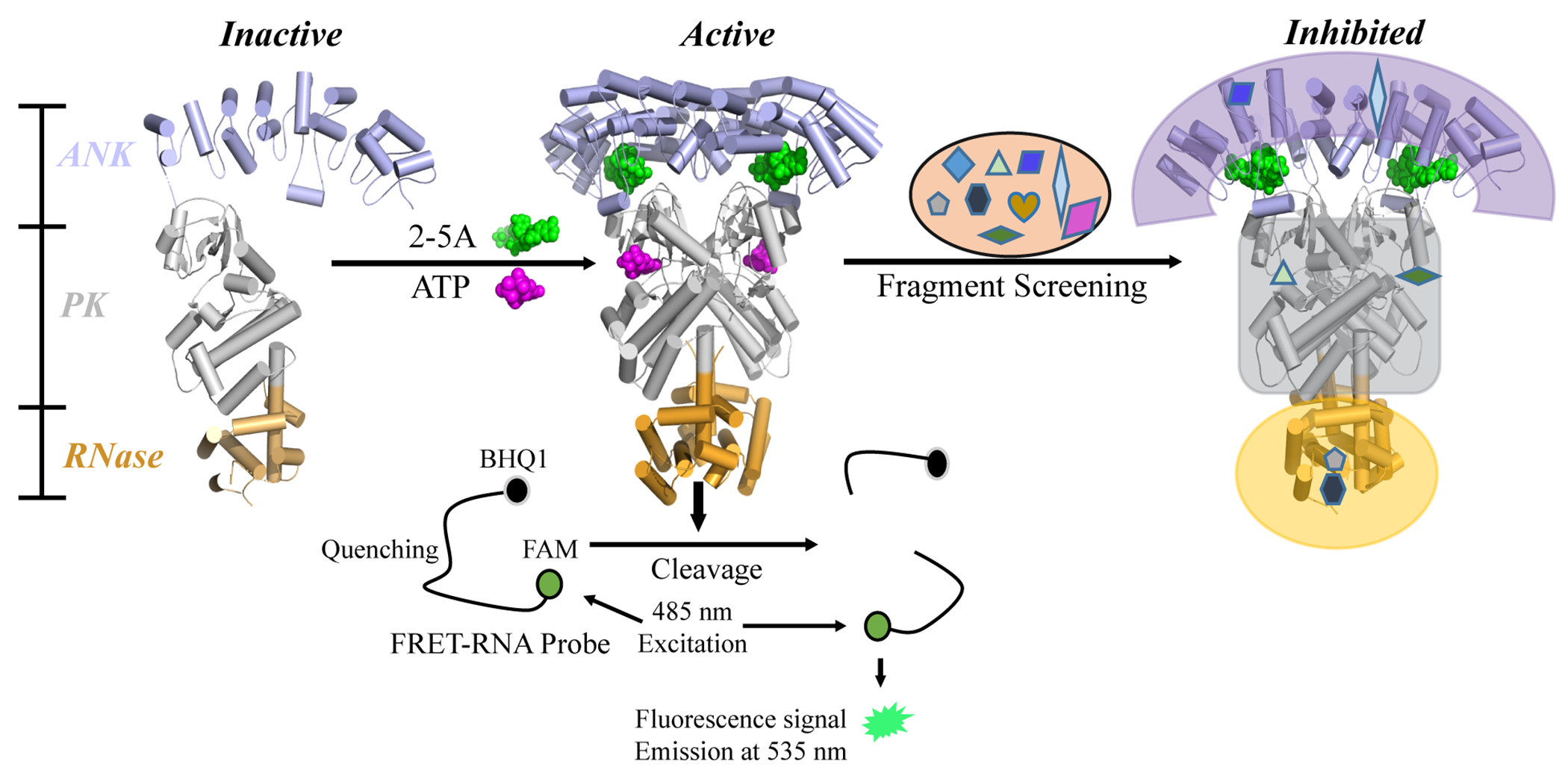
Diagram of RNase L activation and the FRET screening assay. The binding of 2-5A to the ANK domain and the binding of ATP to the PK domain co-operatively induce an active dimer conformation from inactive monomers. Activated RNase L cleaves viral and cellular ssRNA. In the FRET assay, an RNA substrate was labeled with a fluorophore (FAM) and a quencher (BHQ1) at its 5’ and 3’ termini, respectively. Upon substrate cleavage by RNase L, the fluorescence signal of FAM is detected, with excitation at 485 nm and emission at 535 nm. Active fragments were selected for next-stage validation.
