Figure 1.
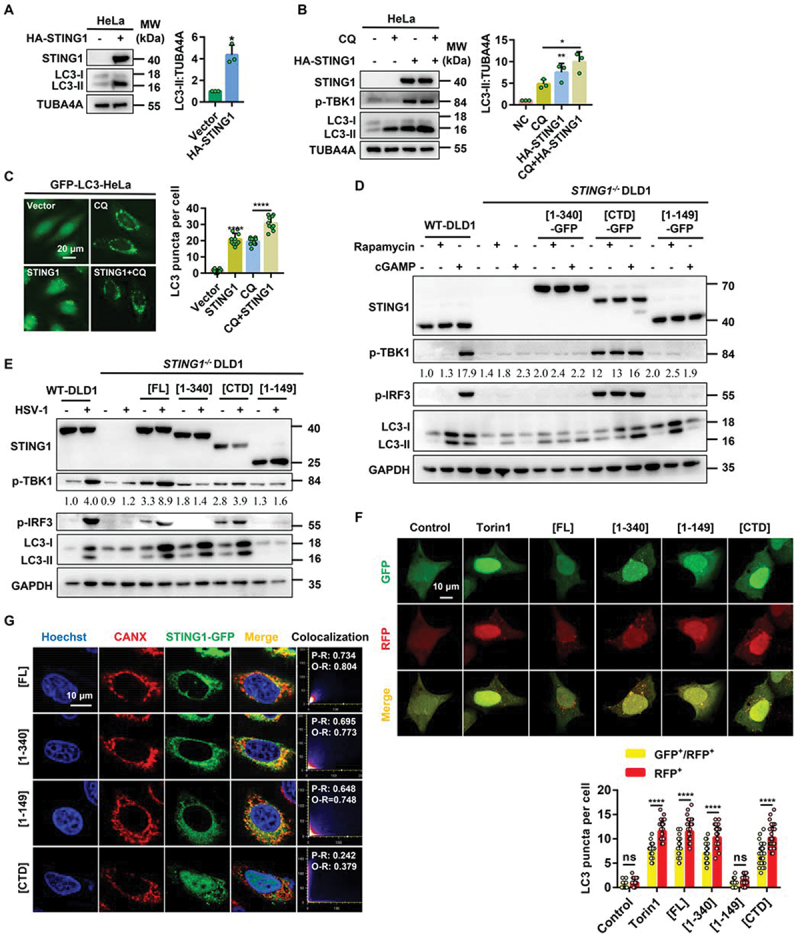
STING1 overexpression and cGAMP induce autophagy. (a) Immunoblot analysis of LC3-ΙΙ levels in HeLa cells that were transfected with vector or HA-STING1 (0.8 µg/mL) for 24 h. (b) Immunoblot analysis of LC3-ΙΙ levels in HeLa cells that were transfected with vector or HA-STING1 with or without CQ treatment at a final concentration of 40 µM. (c) GFP-LC3 puncta formation was analyzed in HeLa cells stably expressing GFP-LC3 that transfected with empty vector or STING1 plasmid with or without CQ (40 µM) treated for 6 h. (d) Western blotting of cell lysates from wild type and STING1-deficient DLD1 cells were transfected with full-length STING1 (STING1[FL]-GFP) or the indicated truncated plasmids labeled with GFP-tag and treated with rapamycin (10 µM) or cGAMP (1 µM) for 6 h. The ratio of p-TBK1:GAPDH was calculated and labeled. (e) Immunoblot analysis of LC3-ΙΙ levels in WT and STING1−/− DLD1 cells that transfected with STING1 or its truncated mutants (STING1 [1-340], STING1[CTD], STING1 [1-149]) with or without HSV-1-GFP virus infected at a multiplicity of infection (MOI) of 5 for 6 h. The ratio of p-TBK1:GAPDH was calculated and labeled. (f) HEK-293A cells stably expressing GFP-RFP-LC3 were treated with 2 μM of torin1 for 6 h or transiently transfected with STING1[FL] or the indicated truncated plasmids. After 24 h expression, cells were harvested and re-seeded on confocal culture dish. The colocalization of GFP and RFP puncta was examined and quantified. Bar: 10 μm. At least 50 cells were counted from each group. (g) WT HeLa cells stably expressing GFP-tagged STING1 or its truncated mutants (STING1 [1-340], STING1[CTD], and STING1 [1-149]) were fixed and stained with marker antibody for ER (CANX [calnexin]) and images were then captured by confocal microscopy. Scale bar: 10 µm. The degree of colocalization for STING1 and CANX was quantified by Pearson’s and overlap coefficient by ImageJ.
*P<0.05, **P<0.01, ***P<0.001, ns, not significant.
