Abstract
Lung cancer remains the leading cause of cancer-related deaths worldwide. Despite the recent advances in cancer therapies, the 5-year survival of non-small cell lung cancer (NSCLC) patients hovers around 20%. Inherent and acquired resistance to therapies (including radiation, chemotherapies, targeted drugs, and combination therapies) has become a significant obstacle in the successful treatment of NSCLC. c-Myc, one of the critical oncoproteins, has been shown to be heavily associated with the malignant cancer phenotype, including rapid proliferation, metastasis, and chemoresistance across multiple cancer types. The c-Myc proto-oncogene is amplified in small cell lung cancers (SCLCs) and overexpressed in over 50% of NSCLCs. c-Myc is known to actively regulate the transcription of cancer stemness genes that are recognized as major contributors to tumor progression and therapeutic resistance; thus, targeting c-Myc either directly or indirectly in mitigation of the cancer stemness phenotype becomes a promising approach for development of a new strategy against drug resistant lung cancers. This review will summarize what is currently known about the mechanisms underlying c-Myc regulation of cancer stemness and its involvement in drug resistance and offer an overview on the current progress and future prospects in therapeutically targeting c-Myc in both SCLC and NSCLC.
Keywords: Cancer dormancy, Cancer stem cells, c-Myc, Drug resistance, Epidermal growth factor receptor, Lung cancer, Non-small cell lung cancer, Small cell lung cancer
Introduction
There are two pathological subtypes of lung cancer. The non-small cell lung cancer (NSCLC) subtype, which includes adenocarcinomas, squamous cell carcinomas, and large cell carcinomas, accounts for 80–85% of all cases. Adenocarcinomas are the most common type of NSCLC and present in the outer parts of the lung. It is less associated with smoking than the other subtypes and more frequently found in women. Though it frequently spreads to the lymph nodes and distant tissues, one third of this type of cancers are often at stage I when diagnosed. Squamous cell carcinomas arise in the central area of the chest in the bronchi and are heavily associated with smoking. This type of cancer originates from the bronchi, similar to the one from epidermal cells, and forms a large cavity within the lung. Large cell carcinomas are a form of undifferentiated carcinoma and are the least common of NSCLC. It can present in any area of the lung and is known to proliferate and metastasize faster than the other subtypes. Small cell lung cancer (SCLC) is the other major pathological subtype that is marked by a very high proliferation rate and strong propensity for metastasis. It is also strongly associated with tobacco smoke exposure. Lung cancer frequently shows no symptoms in its early stages, when it is more easily treated. Advanced disease is characterized by shortness of breath, fatigue, chest pain, and a persistent cough.
Despite being the second most common form of cancer behind breast cancer, lung cancer remains the highest cause of cancer-caused death, accounting for almost 25% of cancer deaths worldwide, more than colon, breast, and prostate cancers combined.1 In 2020, there were a total of 2.21 million new cases of lung cancer and 1.80 million deaths.2 The number of new lung cancer cases has been trending down, but the survival rate still hovers around 20% internationally, despite recent major advances in immunotherapy (immune checkpoint inhibitors that target programmed death-1 [PD-1] and programmed death ligand 1 [PD-L1]) and targeted therapies (inhibitors that target cancer cells harboring specific mutant forms of proliferative signaling proteins such as epidermal growth factor receptor [EGFR], BRAF, and KRAS). The current standard of care involves surgical resection of tumor followed by chemotherapy (usually dual etoposide and platinum) to eliminate residual cancer cells. If the cancer has reached advanced stage and is too large to be removed, combination radiation and chemotherapy are used to first shrink the tumor.3 Immune checkpoint inhibitors are now often administered after four to six cycles of chemotherapy, in addition to any specific inhibitors that would be warranted based on genetic testing.4 Notwithstanding these new therapies, a subset of cancer cells is frequently either inherently resistant or acquires resistance; and often the stage of cancer, graded by its level of differentiation and invasiveness, is correlated to the level of resistance.5
As one of the important oncogenic proteins for cancer growth, c-Myc also plays a crucial role in lung cancer development, progression, and drug resistance. This review will summarize what is currently known about the mechanisms underlying c-Myc regulation of cancer stemness and its involvement in drug resistance and offer an overview on the current progress and future prospects in therapeutically targeting c-Myc in both SCLC and NSCLC.
c-Myc oncoprotein
The c-myc gene was initially discovered 40 years ago as the retroviral v-myc oncogene that was responsible for the transformation of fibroblasts in cell culture.6 Later, it was found that v-myc was not a virus specific gene, instead retrovirally acquired from the mammalian c-myc gene.7,8 Over time, Myc dysregulation in many types of cancers was unveiled, initially by genetic amplifications and chromosomal translocations but eventually through upstream and downstream pathways as well.9 In normal cells, the c-myc gene (located on chromosome 8, band q24.21, with three exons plus two introns) is responsive to various mitogenic and developmental signals. The promoter of the c-myc gene responds to various upstream signaling pathways, such as Wnt/β-catenin, Janus kinase/signal transducer and activator of transcription (JAK/STAT), RAS/RAF/mitogen-activated protein kinase (MAPK), transforming growth factor-β (TGF-β), Notch, and nuclear factor-κB (NF-κB). The protein acts as a transcription factor for the regulation of gene expression involved in a whole host of processes, such as cellular proliferation, metabolism, and differentiation.10 The alterations that occur in cancer, be they chromosomal or non-chromosomal, generally result in a heightened expression of the c-Myc protein (as little as two-fold relative to normal) that plays a significant role driving the unrestrained growth that characterizes oncogenesis.11 This “quantitative” dysregulation contrasts with other oncogenic proteins, such as Ras, Src, and the tumor suppressor p53, whose specific mutations often affect the function of these proteins.12, 13, 14
c-Myc is a member of the superfamily of basic helix-loop-helix leucine zipper (bHLHLZ) DNA binding proteins. It is a global transcriptional regulator with the ability to regulate (either positively or negatively) the expression of 10–15% of the genome, including proteins, both ribosomal and non-ribosomal, and non-coding, regulatory RNAs.15 As a transcription factor, c-Myc directly binds to DNA at E-box domains (CACGTG or similar sequences) as a heterodimer with one of its bHLHLZ interacting partners, Max being the most common. As mentioned previously, the genes under its regulation span a host of processes: cell cycle, differentiation, apoptosis, metabolism, DNA repair, ribosomal biogenesis, protein translation, messenger RNA (mRNA) transcription, cellular migration, angiogenesis, immune response, and stemness.16 In addition to its role as a transcription factor, there have also been reported non-transcriptional roles in DNA synthesis and protein translation.17
c-Myc is a 62 kDa protein with 439 amino acids [Fig. 1]. It is characterized by four conserved domains that are shared by L-Myc and N-Myc. These are denoted as Myc homology boxes (MBs). The N-terminus contains MBI and MBII and is a largely unstructured domain involved in gene transactivation and repression as well as protein regulation. MBI and MBII are located at amino acids (aa) 45–63 and 129–143, respectively. They are largely responsible for the regulation and transcriptional functions of c-Myc. MBI is notable for containing T58 and S62, the phosphorylation of the former being critical for the ubiquitin-proteasomal degradation of c-Myc, whereas the phosphorylation of the latter promoting its stability. While deletion of MBI only partially stifles the oncogenic role of c-Myc, MBII is critical for driving transformation and activating its major target genes.18,19 MBII interacts directly with the large scaffold protein—transformation/transcription domain-associated protein (TRRAP)—that forms a complex with histone acetyl transferases, such as general control non-derepressible 5 (GCN5) and Tip60, and increases acetylation at histone promoters, promoting transcription. c-Myc also binds directly to the histone acetyl transferases p300 and CreB-binding protein (CBP).20 MBII can also interact with other factors, such as positive transcription-elongation factor-b (P-TEFb) that is an RNA polymerase II pause release factor.17
Fig. 1.
Schematic of c-Myc protein. MB domains, calpain cleavage site, and bHLHLZ domain are depicted with corresponding amino acid coordinates. BD: Binding domain; bHLHLZ: Basic helix-loop-helix leucine zipper; MB: Myc homology box; NLS: Nuclear localization signal.
The middle segment of c-Myc contains a region rich in proline, glutamic acid, threonine, and proline residues (PEST), MBIII and MBIV, a calpain cleavage site, as well as a nuclear localization sequence. Less is known about MBIII and MBIV; however, previous reports have shown that they play roles in mediating apoptosis, transformation, transcription repression, and modulating DNA binding.21 Cleavage by the calpain protease has been shown to generate a cytoplasmic form of c-Myc, called Myc-nick (c-Myc-n), which plays a role in promoting cancer cell survival under hypoxia and other stressful conditions.22 The C-terminus contains the bHLHLZ region that dimerizes with the HLHLZ region of Max to form a stable four helix bundle. This heterodimer can specifically interact with DNA by formation of induced fit helices through bonding of the basic regions to residues inside the major grooves of DNA. The Myc–Max interface also serves as a binding region for other factors, such as Miz-1 and Skp2 that influence c-Myc transcription and stability, respectively.23
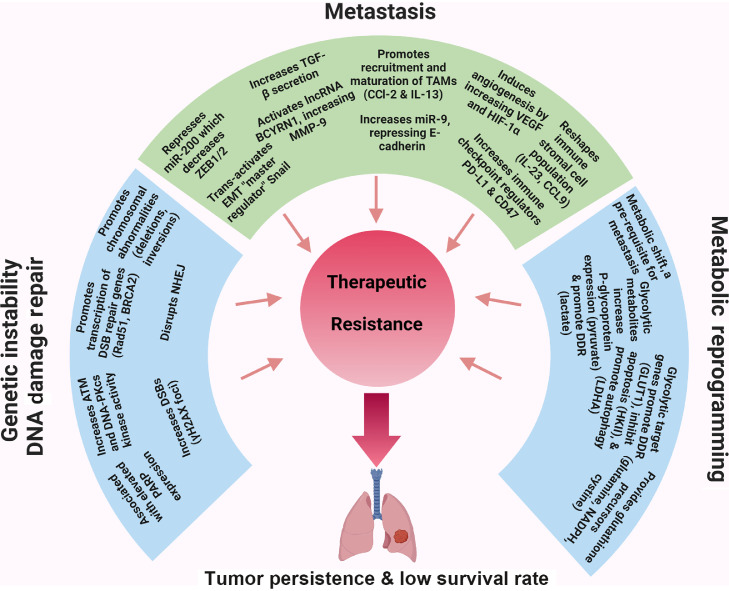
As a master regulator of cell proliferation and survival pathways, c-Myc is implicated in many of the known resistance mechanisms, which will be discussed in the next section. Accordingly, c-Myc protein level has been previously shown to correlate with cancer resistance,24 and there are numerous ongoing trials to target c-Myc directly and indirectly to increase the effectiveness of chemotherapy and other therapeutics.24,25 These, as summarized in Table 1, and future directions will be discussed following a description of c-Myc mediated resistance mechanisms that are highlighted in Fig. 2.
Table 1.
A list of c-Myc inhibitors on clinical trials for potential anti-lung cancer therapies.
| Inhibitor category | Target | Compound | Phase | Trial number |
|---|---|---|---|---|
| Transcriptional | BET | Molibresib, ZEN003694 | Phase I/II and phase II | NCT01587703 and NCT05607108 |
| CDK7 | THZ1, SY 5609 | Preclinical and phase I | N/A, NCT04247126 | |
| CDK9 | SNS032, LY2857785, and AZD4573 | All preclinical | N/A | |
| G-quadruplex | APTO-253 | Phase I | NCT123226 | |
| Translational | mTOR | MLN0128 | Phase I | NCT04250545 |
| eIF4A | Silvestrol, eFT226 | Preclinical and phase I/II | N/A, NCT04092673 | |
| CRI-BPs | Compound 7773 | Preclinical | N/A | |
| Protein stability | USP28 | Compounds 12 and 19 | Preclinical | N/A |
| PLK1 | Volasertib, BI 2536, and onvansertib | Phase II, phase II, and phase II | NCT00824408, NCT00412880, and NCT05450965 | |
| AURKA | Alisertib | Phase II and phase I | NCT01045421 and NCT04085315 | |
| PP2A | FTY720 | Preclinical | N/A | |
| PIN1 | KTP-6566 | Preclinical | NA | |
| Transcription factor activity | Myc–Max heterodimers (two classes) | Small molecule inhibitors (MYCi975) | Preclinical | N/A |
| Large peptide inhibitors (OmoMyc) | Phase I/II | NCT04808362 |
AURKA: Aurora kinase A; CDK: Cyclin dependent protein kinase; BET: Bromodomain and extraterminal; CRI-BPs: Coding region instability-binding proteins; eIF4A: Eukaryotic translation initiation factor 4A; mTOR: Mechanistic target of rapamycin; N/A: Not available; PIN1: Peptidyl-prolyl cis-trans isomerase 1 in human; PLK1: Polo-like kinase 1; PP2A: Protein phosphatase 2A; USP: Ubiquitin specific protease.
Fig. 2.
Mechanisms underlying c-Myc mediated therapeutic resistance. The three general mechanistic aspects, including DNA damage and repair, metastasis, and cancer metabolism, which account for c-Myc-mediated cancer stemness phenotype and therapeutic resistance, are highlighted with examples as described in the review text. ATM: Ataxia-telangiecta-sia mutated; CCL: Chemokine C-C motif ligand; DDR: DNA damage repair; DNA-PKcs: DNA-dependent protein kinase catalytic subunit; DSB: Double strand break; EMT: Epithelial-to-mesenchymal transition; GLUT1: Glucose transporter-1; HIF-1α: Hypoxia inducible factor-1α; HKII: Hexokinase II; IL: Interleukin; LDHA: Lactate dehydrogenase A; lncRNA: Long non-coding RNA; miR: MicroRNA; MMP-9: Matrix metalloproteinase-9; NADPH: Nicotinamide adenine dinucleotide phosphate; PARP: Poly ADP-ribose polymerase; PD-L1: Programmed death ligand 1; TAMs: Tumor-associated macrophages; TGF-β: Tranforming growth factor-β; VEGF: Vascular endothelial growth factor; ZEB: Zinc finger E-box binding homeobox.
c-Myc mediated resistance mechanisms
There are several characteristic features of cancer stem cells that are recognized to contribute to therapeutic resistance, including increased drug efflux transporters, more active DNA repair capacity, increased metastasis, immune escape, altered metabolism, increased autophagy, etc. c-Myc has been shown to mediate these and others, which are detailed in the section below and highlighted in Fig. 2.
Dual role in genetic instability and DNA damage repair
One of the key mechanisms by which c-Myc applies oncogenic stress is through its enabling of uncontrolled cell division. Cell division, though a highly faithful process, is not perfect and generates DNA replication errors that either go uncorrected or are inaccurately corrected, leading to mutations, such as point mutations, gene amplification, and chromosomal translocations. Logically, the higher the mutational burden and genetic instability of a tumor, the higher the chances are of acquiring resistance genes [Fig. 2].26 One of the most important resistance genes is the efflux pump, notably P-glycoprotein, which allows cancer cells to reduce their intracellular concentration of chemotherapy drug.27 As a transcription factor, c-Myc induces expression of a variety of cell cycle, DNA synthesis, and protein translation target genes as well as other factors, such as Cks1 and Cul1, which block cell cycle inhibitors. Active c-Myc has been shown to drive serum starved G0 cells into S phase in the absence of any growth promoting mitogens, one of the hallmarks of cancer.28 In a non-transcriptional role, c-Myc has been found to be an “illegitimate licensing factor” present at the initial sites of DNA replication.29 As such, the increased rate of DNA replication can lead to an excess of replication errors, evidenced by the increase in chromosomal abnormalities (deletions, inversions, and aneuploidy) that occurs following c-Myc induction.30 More lines of evidence come from the increase in double strand breaks (DSBs) that occur following c-Myc overexpression, observed experimentally by an increase in γH2AX foci, a variant of histone H2A protein family and biomarker for DSBs.31 It is also thought that c-Myc can induce DSBs by increasing reactive oxygen species (ROS) as well as by disrupting non-homologous end joining (the major DSB repair mechanism in mammals).32,33 c-Myc also downregulates cell cycle check point proteins, such as bridging integrator 1 (BIN1), which monitor for DNA replication errors and would normally lead to senescence or apoptosis. In addition, c-Myc regulates anti-apoptotic proteins such as B-cell leukemia/lymphoma-2 (Bcl-2) that can prevent the propagation of cells with DSBs.
Paradoxically, following the excessive DNA damage caused by some chemotherapy agents, c-Myc has been shown to promote genomic instability and DSB repair. c-Myc localizes to the promoters of DSB-repair genes, such as Rad51, BRCA2, and DNA-dependent protein kinase catalytic subunit (DNA-PKcs), and promotes their transcription.34 There is also active research on whether c-Myc is essential for the DNA repair process, as there is already some evidence of decrease in kinase activity of ataxia-telangiectasia mutated (ATM) and DNA-PKcs when c-Myc is experimentally reduced.35 Overexpression of c-Myc is also associated with elevated poly ADP-ribose polymerase (PARP) expression, and as such there has been some success in combining chemotherapy with PARP inhibitors.36,37 Finally, c-Myc expression is often elevated in the resistant fraction following cisplatin treatment, as it not only promotes survival through the acquiring of chemo-resistance genes and upregulation of DNA repair proteins, but also has been found to be stabilized in certain cancers by chemo-induced gain or loss of micro-RNAs (miRNAs, such as miR-145) that target c-Myc.38
Role in metastasis
Metastasis accounts for more than 90% of cancer deaths worldwide. Lung cancer (both NSCLC and SCLC) is one of the most lethal forms of cancer due to its high rate of metastasis.39 Primary tumors that gain a metastatic phenotype are highly associated with drug resistance, and thus metastatic pathways have become a major target for anti-cancer drug discovery. Metastasis is both a complex and inefficient process involving cancer cell loss of polarity and subsequent gain of motility, intravasation, circulation, extravasation, seeding, and then repolarization and proliferation in tissue distant from the primary tumor. Previous studies, both in vitro and in vivo, have demonstrated that c-Myc, cooperating with a host of other transcription factors and downstream targets, plays a major role in promoting many of the individual steps of metastasis. This section will go through what is currently known about how c-Myc promotes the different stages of metastasis, both within the tumor cells themselves and the tumor infiltrating cells, which include tumor infiltrating stromal cells, notably cancer-associated fibroblasts (CAFs), and tumor-infiltrating immune cells, comprising many types of innate immune cells, particularly tumor-associated macrophages (TAMs), as well as some types of adaptive immune cells [Fig. 2].
Role in epithelial-to-mesenchymal transition (EMT)
The EMT is a natural, reversible biological process that occurs in a variety of normal biological processes: in embryonic development, organ morphogenesis, wound healing, and tissue fibrosis. It involves an intricate transcriptional program, coinciding with significant epigenetic changes (i.e., major chromatin remodeling), which results in a loss of epithelial characteristics (polarity and cell-to-cell adhesion) and gain of mesenchymal characteristics (non-polarity and cell motility).40 Alterations in specific signaling pathways and cytoskeletal rearrangement are also a necessary component of the EMT process.41 EMT is thought to be at least one way in which cancer cells gain the motility necessary for invasion, although it is clear from in vivo experiments that it is not the only mechanism at play.42 EMT-inducing transcription factors (EMT-TFs) repress the transcription of endothelial genes (notably CDH1 encoding for E-cadherin) that are involved in cellular adhesion, polarity, and cytoskeletal organization and activate the transcription of mesenchymal-associated genes (such as N-cadherin and vimentin). EMT-TFs include SNAIL, slug, zinc finger E-box binding homeobox (ZEB) 1, ZEB2, Twist1, Twist2, E47, and Tcf 3. A network of miRNAs (such as miR-34 and miR-200) and long non-coding RNA (lncRNAs) (such as metastasis associated lung adenocarcinoma transcript 1 [MALAT1] and HOX transcript antisense intergenic RNA [HOTAIR]) are involved in the EMT process as well at a variety of levels: directly targeting the endothelial and mesenchymal proteins themselves, regulating EMT-TFs, and altering the activity of epigenetic remodeler enzymes. Epigenetic remodeling enzymes that are involved in EMT include: DNA methyltransferase 1 (DNMT1), lysine-specific demethylase 1A (LSD1), polycomb repressive complexes 1 and 2 (PRC1/2), suppressor of variegation 3-9 homolog 1 (SUV39H1), SET domain containing protein 8 (SET8), sirtuin 1 (SIRT1), and histone deacetylase 1/2 (HDAC1/2). Regarding drug resistance, the anti-apoptotic proteins Bcl-2 and B-cell lymphoma-extra large (Bcl-XL) have been shown to promote EMT by increasing ROS production.43 Also, the expression of EMT-TFs has been directly linked to drug resistance in lung cancers. Slug was shown to be involved in drug resistance to a traditional EGFR tyrosine kinase inhibitor in NSCLCs.44
Unsurprisingly, c-Myc has been shown to interact with many of these EMT-related factors. Chromatin immunoprecipitation-quantitative polymerase chain reaction (CHIP-qPCR) and luciferase reporter experiments have confirmed c-Myc to directly promote the transcription of SNAIL, the so-called “master regulator” of EMT.45,46 In addition, it has been shown to be indirectly associated with ZEB1/2 transcription.47 c-Myc also associates with epigenetic remodeling enzymes to increase transcription of the EMT-TFs and promote metastasis. Dot1-like (DOT1L), which methylates H3K79 to activate transcription, cooperates with c-Myc/p300 to further unwind the chromatin in the promoter regions of SNAIL and ZEB1/2, by acetylating H3.48 Recently, GATA zinc finger domain containing 2B (GATAD2B), a member of the nucleosome remodeling complex, was shown to directly interact with c-Myc in a KRAS-driven lung cancer to promote EMT-driven metastasis.49 c-Myc directly induces the transcription of miRNAs that promote EMT. In breast cancer, c-Myc directly stimulates miR-9 that in turn downregulates E-cadherin expression.50 Further, c-Myc transcriptionally represses miR-200 that in turn downregulates ZEB1/2, while the latter reduces E-cadherin expression.51 LncRNAs also interact with c-Myc to promote EMT. Actin filament-associated protein1 antisense RNA1 (AFAP1-AS1), which is highly upregulated in many lung cancers, directly binds to a negative regulator of c-Myc, Smad nuclear-interacting protein 1 (SNIP1), preventing its ubiquitination and promoting its stability, thus promoting the transcription of SNAIL and ZEB 1/2.52 c-Myc also interacts with signaling proteins that promote EMT. Proto-oncogene serine/threonine-protein 1 (PIM1) is a serine/threonine kinase that has oncogenic activity in different types of cancer.53 It has been shown to be induced in breast cancer cells by the interleukin (IL)-6-STAT3 signaling pathway and to promote EMT. c-Myc was shown to be a cofactor of PIM1, and knockdown of c-Myc significantly reduced the PIM1-mediated EMT.54 Ubiquitin conjugating enzyme E2 O (UBE2O) is an E2 ubiquitin conjugating enzyme that is known to promote EMT in breast cancer cells. It was recently shown that UBE2O exists in a positive feedback loop with c-Myc through the UBE2O/adenosine 5′-monophosphate (AMP)-activated protein kinase α2 (AMPKα2)/mechanistic target of rapamycin complex 1 (mTORC1)-Myc axis. Many of the CAFs and TAMs within the tumor stroma participate in paracrine signaling with cancer cells by secreting cytokines upregulated by c-Myc, notably TGF-β, which directly promotes EMT within the tumor, with c-Myc being shown to cooperate with SMAD proteins in upregulating EMT master regulator SNAIL.45,55
Role in extracellular matrix (ECM) remodeling
In addition to EMT, there are other major processes that are necessary for successful metastasis, including but not limited to remodeling of the ECM components, angiogenesis, and immune system modulation and evasion. Adhesion to ECM components (such as collagen, elastin, and fibronectin) must be overcome for tumor cell motility and dissemination. c-Myc upregulates a variety of proteases that can cleave these ECM molecules, most notably matrix metalloproteinases (MMPs) and cathepsins.56,57 MMP-9 is the key MMP family member involved in the degradation of collagens and fibronectins to facilitate tumor migration. Though not a direct transcriptional target, c-Myc expression in tumors and TAMs has been shown to drive MMP-9 secretion. Recently, Hu and Lu58 showed the lncRNA brain cytoplasmic RNA 1 (BCYRN1) is activated by c-Myc to promote metastasis in NSCLC by secretion of MMP-9 and MMP-13.58 Pello et al59 used a genetically engineered mouse model to delete c-Myc in myeloid precursor cells and demonstrated that this inhibits the maturation of TAMs, decreases MMP-9 expression, and limits tumor metastasis.59 c-Myc expression has been shown repeatedly to be important for the recruitment and maturation of TAMs, which have many roles in promoting metastasis, including MMP secretion. c-Myc was shown to cooperate with EMT-TF Twist1 to induce chemokine C-C motif ligand 2 (CCL2) and IL-13 secretion by hepatic cell cancer (HCC) cells, which elicit the recruitment and polarization of TAMs. In vivo blockade of these two cytokines drastically reduced TAM infiltration and subsequently abrogated HCC metastasis.60
Role in angiogenesis
Angiogenesis is another key step in the metastatic cascade. Tumors often exist in poorly vascularized microenvironments, lacking access to a blood supply that provides not only the necessary oxygen and nutrients for survival but also the opportunity for cancer cells to intravasate and circulate. c-Myc upregulates both vascular endothelial growth factor (VEGF) and hypoxia inducible factor-1α (HIF-1α), which act in concert to increase the leakiness of surrounding blood vessels and to recruit endothelial cells for the formation of new ones. TAMs predominately occupy the hypoxic areas of tumors, where they secrete VEGF, and are thus thought to be one of the principal mediators of angiogenesis in lung adenocarcinomas.61 There is also evidence of VEGF inducing the recruitment and polarization of TAMs in the presence of certain cytokines, which can further promote the “angiogenic switch” or the continuous growth of vasculature within tumors.62 c-Myc expression in CAFs has been shown to promote the secretion of cytokines that induce the formation of blood vessels by recruiting endothelial cells.63 There are other c-Myc induced proteins and non-coding RNAs that promote angiogenesis. The study of Ciribilli and Borlak64 identified 25 novel target genes of c-Myc in a transgenic mouse model of lung cancer, some of which, such as G-protein couple receptor Adora2B, induced angiogenesis by either inducing VEGF or downregulating negative regulators of HIF-1α.64
Role in immune evasion
Immune evasion involves a series of wholesale changes in the cancer cells and tumor microenvironment to prevent immune clearance of tumor cells. Early clearance of tumor cells must be overcome for successful metastatic invasion, and c-Myc has been linked to many of the immune escape processes. The upregulation of immune checkpoint regulators, such as programmed death ligand 1 (PD-L1) and CD-47, has been linked to c-Myc expression in many cancers, including lymphomas, leukemias, and NSCLCs.65,66 Ras and c-Myc have also been shown to cooperate in promoting lung adenocarcinoma progression by promoting an anti-inflammatory, immunosuppressive environment. It was recently shown that IL-23 and CCL9, both of which are upregulated upon c-Myc deregulation in a genetically engineered mouse model of mutant K-Ras driven lung adenoma, are largely responsible for reshaping the immune stromal cell population. IL-23 rapidly diminishes T-cells, B-cells, and natural killer (NK)-cells; on the other hand, CCL9 potentiated macrophage influx, which promoted angiogenesis through VEGF expression and T-cell exclusion by surface expression of PD-L1.67 IL-23 additionally has been associated with the recruitment of T helper cell 17 (Th-17) lymphocyte, which can convert into immunosuppressive T regulatory cells.68 Also, in lung adenomas, c-Myc has also been shown to downregulate the major histocompatibility complex (MHC) class I, which is an activator of NK-like cells.69
Role in metabolic reprograming
The altered metabolism of rapidly proliferating cancer cells has been recognized as a hallmark of cancer since Warburg's initial proposal in the 1920s.70 This was initially recognized as a means for tumor cells to accumulate biomass precursors by the shuttling of glycolytic intermediates into the pentose phosphate pathway (PPP) but has since been shown to also have a role in drug resistance.71 As c-Myc directly regulates membrane glucose transporters as well as many key glycolytic enzymes, it also mediates this mode of tumor resistance [Fig. 2]. One mechanism of resistance is through the protection of tumor cells from oxidative damage by ROS, which are produced in higher quantities in rapidly proliferating cells. The PPP byproduct nicotinamide adenine dinucleotide phosphate (NADPH) serves as a precursor to glutathione, which can neutralize these harmful ROS by acting as a reducing agent. Also, oxidative phosphorylation (OXPHOS) is a source of ROS species; so, by shifting metabolism toward glycolysis, tumor cells can indirectly become protected by reducing a source of ROS. EMT and its role in conferring drug resistance have already been discussed, but the shift to a glycolytic metabolism has been shown to be a prerequisite for the EMT process and the associated cancer stem cell phenotype.72
Many of the c-Myc regulated glucose transporters and glycolysis enzymes serve non-enzymatic functions in protecting cells from DNA damage and apoptosis that are induced by anti-tumor drugs [Fig. 2]. Glucose transporter-1 (GLUT1) has been shown to be involved indirectly in promoting the DNA damage repair (DDR) pathway by facilitating chromatin remodeling.73 Hexokinase II (HKII) was shown to directly bind to the mitochondrial membrane, inhibiting the release of cytochrome c and subsequent apoptosis that is induced by Bax.74 Pyruvate kinase M2 (PKM2), in addition to promoting the shunt of glycolytic intermediates into the PPP, has been shown to transcriptionally promote the expression of the anti-apoptotic proteins Bcl-xL and Bcl-2.75,76 Autophagy is a catabolic “self-eating” process that cells can utilize to survive under different types of stressors, including apoptosis-inducing chemotherapy and other antitumor drugs. Although c-Myc is involved in upregulating negative regulators of autophagy, such as mechanistic target of rapamycin (mTOR), some of its glycolytic downstream targets can promote autophagy. HKII has been shown to bind to mTOR and inhibit its function, thereby promoting autophagy.77 In addition, lactate dehydrogenase A (LDHA) promotes the formation of the autophagosome.78 Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) has been shown to enhance mitophagy, the specific autophagy of mitochondria, which is a source of ROS and is also necessary for cytochrome-c-mediated apoptosis.79 In addition to the enzymes themselves, the metabolic byproducts of glycolysis, namely pyruvate and lactate, have also been shown to have a role in mediating antitumor drug resistance. Pyruvate was shown to regulate the major drug efflux transporter P-glycoprotein, increasing the efflux of doxorubicin in tumor spheroids.80 Lactate, through its role as an HDAC inhibitor, has been shown to upregulate the DNA repair pathway mediating drug resistance.81 In addition, the acidification of the tumor-surrounding stroma by lactate increases the efficiency of the efflux pumps.82
Another metabolic hallmark of c-Myc upregulation is the increase in glutamine uptake and subsequent glutaminolysis that characterizes rapidly proliferating tumor cells [Fig. 2]. A strong dependence on glutamine by lung cancers, including cisplatin resistant strains of lung cancer, has been described previously.83,84 c-Myc upregulates glutamine transporters, such as alanine-serine-cysteine transporter 2 (ASCT2) and L-type amino acid transporter 1 (LAT1), as well as glutaminases, like glutaminase 1 (GLS1), which are needed to convert glutamine to glutamate for subsequent conversion into α-ketoglutarate, fueling the tricarboxylic acid (TCA) cycle, the intermediates of which are needed for synthesis of purines, pyrimidines, fatty acids, and proteins. When the conversion of glutamate to α-ketoglutarate occurs within the mitochondria, ammonia is also produced, which is a diffusible regulator of autophagy, supporting cell survival under chemotherapy.85 This diffusion can occur not only between tumor cells but also with tumor-associated stromal cells such as CAFs and TAMs. Indeed, in K-Ras driven lung cancers, it was shown that cancer cells lacking glutamine could derive this metabolic precursor from surrounding stroma.86 Glutamine-derived glutamate is also critical for redox control in tumors. Glutamate is a component of reduced glutathione, a tripeptide of glutamate, cysteine, and glycine, which is the major scavenger of oxidative species, namely ROS, that are induced by DNA damage effected by antitumor drugs. c-Myc also upregulates the glutamate-cystine antiporter solute carrier family 7 member 11 (SLC7A11), which is responsible for the import of cystine that is converted into cysteine in the cell. Much the same as the glycolytic switch acting a prerequisite for EMT, silencing of GLS1 has been shown to impair metastasis in lung cancers.87
Therapeutic targeting of c-Myc in NSCLC and SCLC
Historically, c-Myc has been considered an “undruggable” target. This is due to it being a transcription factor, featuring significant structural disorder and lacking a specific inhibition site, as a kinase would have in its enzymatic active site, for example. Also, c-Myc resides predominately in the nucleus, making monoclonal antibody therapy unfeasible. Targeting c-Myc through other means, such as targeting c-Myc transcription, c-Myc translation, and c-Myc stability, as well as c-Myc transcriptional activity, has shown promise in treating lung cancer, with several clinical trials completed and ongoing [Table 1].
Transcriptional inhibitors
Bromodomain and extraterminal (BET) family member bromodomain-containing protein 4 (BRD4) is a transcriptional regulator that recruits the P-TEFb to the c-terminus of RNA polymerase for its phosphorylation and subsequent release from a gene's promoter, thus promoting transcription. The Myc oncogene family is under the regulation of BRD4, and there have been numerous in vitro and in vivo studies showing the effectiveness of BET inhibitor (BETi) against Myc-driven cancers.88, 89, 90, 91, 92 Recently, a phase 1 and 2 clinical trial was completed to study the optimal dosing for the BETi molibresib in patients with solid tumors, including NSCLCs and SCLCs.93 There is also an ongoing clinical trial examining another specific BETi in advanced squamous cell lung carcinomas (ClinicalTrials.gov: NCT05607108). Some evidence of BETi resistance in K-Ras driven NSCLC has recently been found due to concurrent activation of mTOR upon BETi treatment, but combination therapy with mTOR inhibitor was shown to improve sensitivity to BETi.94
A feature of Myc dysregulated cancers is the attainment of super-enhancers as distal cis-regulatory elements that act to significantly amplify c-Myc transcription.95 Super-enhancers are populated by an abundance of common transcription factors but transcriptional activators unique to this area are the less common cyclin dependent kinases CDK7 and CDK9. For this reason, small molecule inhibitors to these kinases have been developed to target super enhancer regulatory elements and the proto-oncogenes under their transcriptional control. The covalent CDK7 inhibitor THZ1 was found to be highly effective against SCLC in a combination in vivo/in vitro study by suppressing the expression of proto-oncogenes such as c-Myc, SRY-box transcription factor 2 (SOX2), and nuclear factor I B (NFIB).96 Three different CDK9 inhibitors were shown to be effective against lung cancer cell lines and organoids with mutations in K-Ras or EGFR as well as osimertinib and AMG510 resistant lung adenocarcinoma cells.97 c-Myc protein levels were not found to be affected but other proteins associated with promoting the cancer stemness state, myeloid cell leukemia 1 (Mcl-1), Sox2, and Sox9, were found to be decreased. There is currently a phase 1 clinical trial of a selective CDK-7 inhibitor, SY 5609, underway to study its effectiveness against advanced stage solid tumors, including SCLC, in adults (ClinicalTrials.gov: NCT04247126).
An alternative approach to targeting the Myc transcriptional machinery is to stabilize secondary structures in the Myc promoter, which must be unwound first prior to transcription. For proto-oncogenes such as c-Myc, guanine-rich DNA sequences in the proximal promoter region fold into four-stranded secondary structures known as G-quadruplexes, and these three-dimensional structures present binding sites for small molecules.98 APTO-253 is a small molecule inhibitor that has been shown to inhibit c-Myc expression by stabilizing G-quadruplexes in DNA.99 A phase 1 trial demonstrated the anti-tumor activity of this compound against metastatic solid tumors, including NSCLC, as well as good tolerability at the requisite dosage.100 There have been several more recent studies characterizing novel G-quadruplex stabilizing small molecules in vitro due to their promise as potent inhibitors of oncogenes such as c-Myc and K-Ras.101, 102, 103 DEAD-box helicase 5 (DDX5) is a protein from the family of double-stranded RNA helicases, which has been found to be overexpressed in many types of cancers, including lung, and has been shown to unwind the G-quadruplex in the Myc gene promoter.104 Thus, compounds that inhibit this protein could also present a promising area of research for reducing c-Myc transcription in cancers that show high DDX5 expression.
Translational inhibitors
c-Myc mRNA has some unique features that allow for its pharmacological inhibition. Its 5′ untranslated regions (UTR) contains complex secondary structures, namely G-quadruplexes, which requires the eukaryotic initiation factor 4F (eIF4F) complex for its successful translation.105 mTOR is a serine/threonine kinase that serves as the catalytic subunit of the mTORC1 protein complex, which phosphorylates its two major effector proteins S6 kinase beta-1 (S6K1) and eukaryotic initiation factor 4E-binding protein 1 (4EBP1), both of which facilitate eIF4F-mediated translation. A major effector protein of SK61 is eukaryotic translation initiation factor 4B (eIF4B), a member of the eIF4F complex, which aids eukaryotic translation initiation factor 4A (eIF4A), a helicase that unwinds the mRNA secondary structure prior to translation initiation. Also, 4EBP1 blocks the binding of eukaryotic initiation factor 4G (eIF4G) to eukaryotic initiation factor 4E (eIF4E), but this negative regulation is attenuated when 4EBP1 is phosphorylated by mTORC1, allowing for the initiation of translation. Thus, an indirect strategy to target c-Myc translation is to pharmacologically inhibit the mTOR kinase or one of the mTORC1 effector proteins.
Traditional inhibitors of mTOR, rapamycin and its analogs, have proven to be clinically unfeasible due to issues with toxicity and stability. Next generation inhibitors of mTOR, such as MLN0128, have been extensively studied recently both in vivo, showing tumor reduction in an Eμ-Myc-driven transgenic mouse model, and in phase 1 and 2 clinical trials, with promising results for many types of advanced stage solid tumors.106, 107, 108, 109 While studies for lung cancer are lacking, there is a clinical trial ongoing examining the efficacy of this drug against NSCLC (ClinicalTrials.gov: NCT04250545). MLN0128 effectively inhibits mTOR phosphorylation of 4EBP1, allowing eIF4G binding to eIF4E; however, there are eIF4E-independent mechanisms for c-Myc mRNA translation initiation, and so treatment with MLN0128 may fail to sufficiently inhibit c-Myc translation in some cancers.110 Inhibiting eIF4A directly can overcome this pitfall because unwinding the 5′UTR G-quadruplexes is indispensable for c-Myc translation. Two inhibitors of eIF4A currently under evaluation are silvestrol and eFT226. Silvestrol has been evaluated in vitro and in vivo, showing potent reduction in c-Myc mRNA translation and tumor size, respectively.111,112 eFT226 is also being studied and is currently the subject of a major phase 1–2 clinical trial evaluating the drug against advanced stage solid tumors, including NSCLC (ClinicalTrials.gov: NCT04092673).
Targeting the stability of c-Myc mRNA is a newer research area that is being explored. There is a family of proteins that bind to specific sequences of the 3′UTR, called cytoplasmic polyadenylation elements (CPEs), which act to destabilize mRNA. These are so-called cytoplasmic polyadenylation binding (CPEB) binding proteins. A very recent study that utilized gene ontology and enrichment analysis in examining miRNA sequencing data from NSCLC tumors showed that CPEB3 was a significantly downregulated protein compared to healthy lung.113 Identifying strategies to reactivate this protein thus could be a promising strategy for targeting c-Myc in lung cancer. In addition to proteins that bind the 3′UTR, there are proteins that bind to coding sequences of mRNA, called coding region instability-binding proteins (CRI-BPs), which act to protect mRNA from endonuclease activity. Insulin-like growth factor 2 mRNA binding protein (Igf2bp) was recently identified as a CRI-BP regulating c-Myc RNA stability using a mouse model of lung adenocarcinoma, in which expression of this protein enhanced tumor progression and metastasis.114 There are ongoing in vitro cell studies to identify small molecule inhibitors of this protein in lung and other cancers such as melanoma. Recently, a small molecule inhibitor was isolated and subsequently shown to decrease the mRNA level of not only c-Myc but also K-Ras in lung cancer cells.115 This is another area of research that may prove to be fruitful in the future for targeting c-Myc and its associated oncogenes in lung cancers.
Targeting proteins that stabilize c-Myc
The stability of c-Myc (a short-lived protein with a 20-min half-life) is governed by phosphorylation at two residues on its N-terminus. Phosphorylation at S62 by several kinases promotes c-Myc stability, whereas phosphorylation at T58 by glycogen synthase kinase 3 (GSK3) promotes its degradation by the ubiquitin-proteosome system (UPS). K-Ras is upstream of extracellular signal-regulated kinase (ERK), one of the kinases that phosphorylate S62, and, in addition, K-Ras activates phosphoinositide 3-kinase (PI3K)/Akt that phosphorylates and inactivates GSK3. Thus, the frequent activating mutations in K-Ras, which are observed in lung cancer, can be easily tied to the overactivation of c-Myc that is frequently found in the same tumors, and explains the efficacy that has been observed in targeting c-Myc in K-Ras driven lung tumors.116, 117, 118 Another protein that is important in regulating the turnover of c-Myc is protein phosphatase 2A (PP2A), which must dephosphorylate S62 after T58 phosphorylation for UPS-mediated degradation to occur. The major E3 ligase that recognizes and binds to phosphorylated T58 is the S-phase kinase associated protein 1 (SKP1)-cullin-1-F-box and WD repeat domain-containing 7 ubiquitin ligase complex (SCF Fbw7), which leads to degradation of c-Myc by the 26S proteosome. There are also several other kinases, deubiquitinases, and c-Myc interacting proteins that are involved in this process that can be pharmacologically targeted to decrease c-Myc protein stability in lung cancer. A summary of these is described below and shown in Table 1.
Deubiquitinases such as ubiquitin specific protease (USP) 28, USP36, and USP37 antagonize E3 ligase activity and stabilize c-Myc. A number of these have been found to be overexpressed in lung cancer.119, 120, 121 Specific inhibitors to USP28 have been recently developed and shown to be effective against lung cancer cell lines as well as in an ex vivo murine lung slice culture system.122,123 Polo-like kinase 1 (PLK1) is serine-threonine kinase involved in the cell cycle, which phosphorylates c-Myc and contributes to its stability. PLK1 has also been found to be overexpressed in lung cancers and associated with poor prognosis.124,125 Inhibitors for PLK1 have been developed and tested in phase II clinical trials. Volasertib was tested in patients with metastatic NSCLC and BI 2536 was examined in patients with relapsed SCLC, both with minimal efficacy, which unfortunately has been the trend for this class of inhibitor with lung cancers.126, 127, 128 New inhibitors have continued to be developed, and there is an ongoing clinical trial with a novel PLK1 inhibitor, onvansertib, in patients with chemo-resistant SCLC (ClinicalTrials.gov: NCT05450965). Aurora kinase A (AURKA) is another cell-cycle related kinase that stabilizes c-Myc and found to be overexpressed in many types of cancers, including lung.129 Alisertib (MLN8237) is a selective inhibitor against AURKA and has shown modest efficacy against lung cancer in clinical trials. In a phase II study, MLN8237 as a monotherapy treatment yielded a 21% objective response rate in 48 patients with SCLC.130 More recently, alisertib was combined with EGFR tyrosine kinase inhibitor osimertinib in a phase I clinical trial, with preliminary results showing that alisertib may improve disease control in patients with lung adenocarcinomas resistant to osimertinib monotherapy.131 PP2A, as previously mentioned, promotes c-Myc degradation. There are two endogenous PP2A inhibitors, su(var)3-9, enhancer of zeste, trithorax (SET)/inhibitor 2 of PP2A and cellular inhibitor of PP2A (CIP2A), which are overexpressed in several types of tumors, including lung.132 FTY720, a SET inhibitor, was shown to increase cell death in A549 cells and xenografts.133 PIN1 was shown to mediate c-Myc-DNA binding,134 although it was previously shown to potentially mediate PP2A-facilitated c-Myc degradation.135 Thus, a covalent small-molecule inhibitor of PIN1 was recently developed, which was shown to selectively target cancer cells in vitro as well as in a mouse model of lung tumorigenesis.136
Targeting c-Myc transcriptional activity
For c-Myc to act as a transcription factor, it must form a heterodimer with one of its bHLHLZ binding partners, notably Max. Several labs have identified small molecules in screens, which are able to block the Myc–Max interaction.137, 138, 139 These compounds, such as MYCi975, have been tested in several cancer cell lines, including lung, with half maximal inhibitory concentration (IC50) values close to 10 μmol/L or less. Several have also been studied in Myc-driven mouse models and showed anti-tumor activity. More promising is the recent success of OmoMyc, a 90 amino acid mini-protein, which mimics the bHLHLZ domain of c-Myc with four key mutations, which allows OmoMyc to bind directly to c-Myc or Max. It can form heterodimers with c-Myc or Max, in addition to forming homodimers with itself, which abrogates c-Myc transcriptional activity, essentially acting as a dominant negative form of c-Myc.140 In a paper published by Peptomyc,141 the company founded by OmoMyc's inventor, the researchers show that purified OmoMyc effectively penetrates cancer cells and blocks c-Myc transcriptional activity. In contrast to standard of care therapies, OmoMyc only targets cancer cells while sparing normal cells due to the low level of endogenous c-Myc in differentiated cells. These preclinical data along with the effectiveness of OmoMyc in various models of NSCLC with different mutational backgrounds gave rise to a phase I clinical trial of OmoMyc in 2020, which enrolled 22 patients with advanced solid tumors, including NSCLC, showing a halt of tumor growth in 8 of the patients with minimal side effects.118,142 Based on these results, this version of OmoMyc (OMO-103) is rapidly proceeding to a phase II trial.
Conclusions and perspective
Herein, we have described the mechanisms by which c-Myc promotes cancer therapeutic resistance, acting to promote a cancer stemness phenotype, in the realms of DNA damage and repair, EMT and metastasis, and cancer metabolism [Fig. 2]. As most subtypes of lung cancer remain notoriously difficult to treat, c-Myc remains an attractive target. Due to the issues described herein, it was long considered undruggable, as were many proteins before it, but considerable progress has been made to indirectly target it, with many of the drugs mentioned having completed clinical trials showing a direct anti-cancer effect [Table 1]. Going forward, efforts to target c-Myc more directly with peptides, like OmoMyc, will no doubt gain prominence, as many of the more indirect methods will suffer from off-target effects. Development of drug resistance still remains a concern, but this can also be assuaged by more directly targeting c-Myc, considering its central role in regulating many different pathways that promote tumorigenesis.
Conflicts of interest
None.
Acknowledgments
H.L. was supported by NIH-NCI grants R01CA234605, R21CA272890, and U01CA252965, as well as the Reynolds and Ryan Families Endowed Chair fund and the Ladies Leukemia League fund. The figures and table were created using BioRender.com.
Edited by: Peifang Wei
References
- 1.Lung Cancer Statistics | How common is lung cancer? 2023. Available from: https://www.cancer.org/cancer/lung-cancer/about/key-statistics.html. [Accessed on February 28, 2023].
- 2.CHEST. World Lung Cancer Day Fact Sheet – American College of Chest Physicians. 2023. Available from: https://www.chestnet.org/Newsroom/CHEST-News/2021/07/World-Lung-Cancer-Day-Fact-Sheet. [Accessed on April 19, 2023].
- 3.Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet. 2019;394:1929–1939. doi: 10.1016/S0140-6736(19)32222-6. [DOI] [PubMed] [Google Scholar]
- 4.Lung Cancer Treatment by Stage | Treating SCLC by Stage. 2023. https://www.cancer.org/cancer/lung-cancer/treating-small-cell/by-stage.html. [Accessed on February 28, 2023].
- 5.Tulpule A, Bivona TG. Acquired resistance in lung cancer. Annu Rev Cancer Biol. 2020;4:279–297. doi: 10.1146/annurev-cancerbio-030419. [DOI] [Google Scholar]
- 6.Sheiness D, Fanshier L, Bishop JM. Identification of nucleotide sequences which may encode the oncogenic capacity of avian retrovirus MC29. J Virol. 1978;28:600–610. doi: 10.1128/JVI.28.2.600-610.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roussel M, Saule S, Lagrou C, et al. Three new types of viral oncogene of cellular origin specific for haematopoietic cell transformation. Nature. 1979;281:452–455. doi: 10.1038/281452a0. [DOI] [PubMed] [Google Scholar]
- 8.Sheiness D, Bishop JM. DNA and RNA from uninfected vertebrate cells contain nucleotide sequences related to the putative transforming gene of avian myelocytomatosis virus. J Virol. 1979;31:514–521. doi: 10.1128/JVI.31.2.514-521.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer N, Penn LZ. Reflecting on 25 years with Myc. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/NRC2231. [DOI] [PubMed] [Google Scholar]
- 10.Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-Myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. Pt 2. [DOI] [PubMed] [Google Scholar]
- 11.Murphy DJ, Junttila MR, Pouyet L, et al. Distinct thresholds govern Myc's biological output in vivo. Cancer Cell. 2008;14:447–457. doi: 10.1016/j.ccr.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sen B, Johnson FM. Regulation of Src family kinases in human cancers. J Signal Transduct. 2011;2011 doi: 10.1155/2011/865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burge RA, Hobbs GA. Not all RAS mutations are equal: a detailed review of the functional diversity of RAS hot spot mutations. Adv Cancer Res. 2022;153:29–61. doi: 10.1016/bs.acr.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhou X, Hao Q, Lu H. Mutant p53 in cancer therapy-the barrier or the path. J Mol Cell Biol. 2019;11:293–305. doi: 10.1093/jmcb/mjy072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105:670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Cole MD, Cowling VH. Transcription-independent functions of Myc: regulation of translation and DNA replication. Nat Rev Mol Cell Biol. 2008;9:810–815. doi: 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conzen SD, Gottlob K, Kandel ES, et al. Induction of cell cycle progression and acceleration of apoptosis are two separable functions of c-Myc: transrepression correlates with acceleration of apoptosis. Mol Cell Biol. 2000;20:6008–6018. doi: 10.1128/MCB.20.16.6008-6018.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oster SK, Ho CS, Soucie EL, Penn LZ. The Myc oncogene: MarvelouslY complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 20.Vervoorts J, Lüscher-Firzlaff JM, Rottmann S, et al. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baluapuri A, Wolf E, Eilers M. Target gene-independent functions of MYC oncoproteins. Nat Rev Mol Cell Biol. 2020;21:255–267. doi: 10.1038/s41580-020-0215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conacci-Sorrell M, Ngouenet C, Anderson S, Brabletz T, Eisenman RN. Stress-induced cleavage of Myc promotes cancer cell survival. Genes Dev. 2014;28:689–707. doi: 10.1101/gad.231894.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Zhang J, Yin J, et al. Alternative approaches to target Myc for cancer treatment. Signal Transduct Target Ther. 2021;6:117. doi: 10.1038/s41392-021-00500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chanvorachote P, Sriratanasak N, Nonpanya N. C-myc contributes to malignancy of lung cancer: a potential anticancer drug target. Anticancer Res. 2020;40:609–618. doi: 10.21873/anticanres.13990. [DOI] [PubMed] [Google Scholar]
- 26.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 27.Seelig A. P-glycoprotein: One mechanism, many tasks and the consequences for pharmacotherapy of cancers. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.576559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermeking H, Wolf DA, Kohlhuber F, et al. Role of c-Myc in simian virus 40 large tumor antigen-induced DNA synthesis in quiescent 3T3-L1 mouse fibroblasts. Proc Natl Acad Sci USA. 1994;91:10412–10416. doi: 10.1073/pnas.91.22.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Kuzyk A, Mai S. c-Myc-induced genomic instability. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.A014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW. Defective double-strand DNA break repair and chromosomal translocations by Myc overexpression. Proc Natl Acad Sci USA. 2003;100:9974–9979. doi: 10.1073/pnas.1732638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vafa O, Wade M, Kern S, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/S1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Owonikoko TK, Sun SY, et al. c-Myc suppression of DNA double-strand break repair. Neoplasia. 2012;14:1190–1202. doi: 10.1593/neo.121258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumari A, Folk WP, Sakamuro D. The dual roles of Myc in genomic instability and cancer chemoresistance. Genes. 2017;8:158. doi: 10.3390/genes8060158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cui F, Fan R, Chen Q, et al. The involvement of c-Myc in the DNA double-strand break repair via regulating radiation-induced phosphorylation of ATM and DNA-PKcs activity. Mol Cell Biochem. 2015;406:43–51. doi: 10.1007/s11010-015-2422-2. [DOI] [PubMed] [Google Scholar]
- 36.Lok BH, Gardner EE, Schneeberger VE, et al. PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin Cancer Res. 2017;23:523–535. doi: 10.1158/1078-0432.CCR-16-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyndiah S, Tanida S, Ahmed KM, Cassimere EK, Choe C, Sakamuro D. c-MYC suppresses BIN1 to release poly(ADP-ribose) polymerase 1: a mechanism by which cancer cells acquire cisplatin resistance. Sci Signal. 2011;4:ra19. doi: 10.1126/scisignal.2001556. [DOI] [PubMed] [Google Scholar]
- 38.Sheng Q, Zhang Y, Wang Z, Ding J, Song Y, Zhao W. Cisplatin-mediated down-regulation of miR-145 contributes to up-regulation of PD-L1 via the c-Myc transcription factor in cisplatin-resistant ovarian carcinoma cells. Clin Exp Immunol. 2020;200:45–52. doi: 10.1111/cei.13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rapp UR, Korn C, Ceteci F, et al. MYC is a metastasis gene for non-small-cell lung cancer. PLoS One. 2009;4:e6029. doi: 10.1371/journal.pone.0006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JY, Kong G. Roles and epigenetic regulation of epithelial-mesenchymal transition and its transcription factors in cancer initiation and progression. Cell Mol Life Sci. 2016;73:4643–4660. doi: 10.1007/s00018-016-2313-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Datta A, Deng S, Gopal V, et al. Cytoskeletal dynamics in epithelial-mesenchymal transition: insights into therapeutic targets for cancer metastasis. Cancers. 2021;13:1882. doi: 10.3390/cancers13081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi X, Zhang L, Lu X. New insights into the epithelial-to-mesenchymal transition in cancer. Trends Pharmacol Sci. 2016;37:246–248. doi: 10.1016/J.tips.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Kim EM, Jung CH, Song JY, Park JK, Um HD. Pro-apoptotic Bax promotes mesenchymal-epithelial transition by binding to respiratory complex-I and antagonizing the malignant actions of pro-survival Bcl-2 proteins. Cancer Lett. 2018;424:127–135. doi: 10.1016/j.canlet.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 44.Chang TH, Tsai MF, Su KY, et al. Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. Am J Respir Crit Care Med. 2012;183:1071–1079. doi: 10.1164/RCCM.201009-1440OC. [DOI] [PubMed] [Google Scholar]
- 45.Smith AP, Verrecchia A, Fagà G, et al. A positive role for Myc in TGFbeta-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene. 2009;28:422–430. doi: 10.1038/onc.2008.395. [DOI] [PubMed] [Google Scholar]
- 46.Cho KB, Cho MK, Lee WY, Kang KW. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett. 2010;293:230–239. doi: 10.1016/j.canlet.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 47.Larsen JE, Nathan V, Osborne JK, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219–3235. doi: 10.1172/JCI76725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho MH, Park JH, Choi HJ, et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nature Commun. 2015;6:7821. doi: 10.1038/ncomms8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grzeskowiak CL, Kundu ST, Mo X, et al. In vivo screening identifies GATAD2B as a metastasis driver in KRAS-driven lung cancer. Nat Commun. 2018;9:2732. doi: 10.1038/s41467-018-04572-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu M, Zhu H, Yang S, Wang Z, Bai J, Xu N. c-Myc suppressed E-cadherin through miR-9 at the post-transcriptional level. Cell Biol Int. 2013;37:197–202. doi: 10.1002/cbin.10039. [DOI] [PubMed] [Google Scholar]
- 51.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong Y, Yang L, Xiong F, et al. Long non-coding RNA AFAP1-AS1 accelerates lung cancer cells migration and invasion by interacting with SNIP1 to upregulate c-Myc. Signal Transduct Target Ther. 2021;6:240. doi: 10.1038/s41392-021-00562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cao L, Wang F, Li S, Wang X, Huang D, Jiang R. PIM1 kinase promotes cell proliferation, metastasis and tumor growth of lung adenocarcinoma by potentiating the c-MET signaling pathway. Cancer Lett. 2019;444:116–126. doi: 10.1016/j.canlet.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 54.Gao X, Liu X, Lu Y, et al. PIM1 is responsible for IL-6-induced breast cancer cell EMT and stemness via c-Myc activation. Breast Cancer. 2019;26:663–671. doi: 10.1007/s12282-019-00966-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calon A, Tauriello DV, Batlle E. TGF-beta in CAF-mediated tumor growth and metastasis. Semin Cancer Biol. 2014;25:15–22. doi: 10.1016/j.semcancer.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 56.Pello OM, De Pizzol M, Mirolo M, et al. Role of c-Myc in alternative activation of human macrophages and tumor-associated macrophage biology. Blood. 2012;119:411–421. doi: 10.1182/blood-2011-02-339911. [DOI] [PubMed] [Google Scholar]
- 57.Brindle NR, Joyce JA, Rostker F, et al. Deficiency for the cysteine protease cathepsin L impairs Myc-induced tumorigenesis in a mouse model of pancreatic neuroendocrine cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu T, Lu YR. BCYRN1, a c-Myc-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 2015;15:36. doi: 10.1186/s12935-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pello OM, Chèvre R, Laoui D, et al. In vivo inhibition of c-MYC in myeloid cells impairs tumor-associated macrophage maturation and pro-tumoral activities. PLoS One. 2012;7:e45399. doi: 10.1371/journal.pone.0045399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dhanasekaran R, Baylot V, Kim M, et al. MYC and twist1 cooperate to drive metastasis by eliciting crosstalk between cancer and innate immunity. Elife. 2020;9:e50731. doi: 10.7554/eLife.50731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Riabov V, Gudima A, Wang N, Mickley A, Orekhov A, Kzhyshkowska J. Role of tumor associated macrophages in tumor angiogenesis and lymphangiogenesis. Front Physiol. 2014;5:75. doi: 10.3389/fphys.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wheeler KC, Jena MK, Pradhan BS, et al. VEGF may contribute to macrophage recruitment and M2 polarization in the decidua. PLoS One. 2018;13 doi: 10.1371/journal.pone.0191040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mundim FGL, Pasini FS, Brentani MM, Soares FA, Nonogaki S, Waitzberg AFL. MYC is expressed in the stromal and epithelial cells of primary breast carcinoma and paired nodal metastases. Mol Clin Oncol. 2015;3:506–514. doi: 10.3892/mco.2015.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ciribilli Y, Borlak J. Oncogenomics of c-Myc transgenic mice reveal novel regulators of extracellular signaling, angiogenesis and invasion with clinical significance for human lung adenocarcinoma. Oncotarget. 2017;8:101808–101831. doi: 10.18632/oncotarget.21981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim EY, Kim A, Kim SK, Chang YS. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer. 2017;110:63–67. doi: 10.1016/j.lungcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 66.Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kortlever RM, Sodir NM, Wilson CH, et al. Myc cooperates with ras by programming inflammation and immune suppression. Cell. 2017;171:1301–1315. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; e14. 10.1016/j.cell.2017.11.013.
- 68.Gomez-Rodriguez J, Wohlfert EA, Handon R, et al. Itk-mediated integration of T cell receptor and cytokine signaling regulates the balance between Th17 and regulatory T cells. J Exp Med. 2014;211:529–543. doi: 10.1084/jem.20131459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16:7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 70.WEINHOUSE S. On respiratory impairment in cancer cells. Science. 1956;124:267–269. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 71.Marcucci F, Rumio C. Glycolysis-induced drug resistance in tumors – a response to danger signals? Neoplasia. 2021;23:234–245. doi: 10.1016/j.neo.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee SY, Ju MK, Jeon HM, et al. Oncogenic metabolism acts as a prerequisite step for induction of cancer metastasis and cancer stem cell phenotype. Oxid Med Cell Longev. 2018;2018 doi: 10.1155/2018/1027453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim J, Xu S, Xiong L, Yu L, Fu X, Xu Y. SALL4 promotes glycolysis and chromatin remodeling via modulating HP1α-Glut1 pathway. Oncogene. 2017;36:6472–6479. doi: 10.1038/onc.2017.265. [DOI] [PubMed] [Google Scholar]
- 74.Pastorino JG, Shulga N, Hoek JB. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J Biol Chem. 2002;277:7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- 75.Kwon OH, Kang TW, Kim JH, et al. Pyruvate kinase M2 promotes the growth of gastric cancer cells via regulation of Bcl-xL expression at transcriptional level. Biochem Biophys Res Commun. 2012;423:38–44. doi: 10.1016/j.bbrc.2012.05.063. [DOI] [PubMed] [Google Scholar]
- 76.Lin Y, Zhai H, Ouyang Y, et al. Knockdown of PKM2 enhances radiosensitivity of cervical cancer cells. Cancer Cell Int. 2019;19:129. doi: 10.1186/s12935-019-0845-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu X, Miao W, Huang M, Li L, Dai X, Wang Y. Elevated hexokinase II expression confers acquired resistance to 4-hydroxytamoxifen in breast cancer cells. Mol Cell Proteomics. 2019;18:2273–2284. doi: 10.1074/mcp.RA119.001576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Das CK, Parekh A, Parida PK, Bhutia SK, Mandal M. Lactate dehydrogenase a regulates autophagy and tamoxifen resistance in breast cancer. Biochim Biophys Acta Mol Cell Res. 2019;1866:1004–1018. doi: 10.1016/j.bbamcr.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Colell A, Ricci JE, Tait S, et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 80.Wartenberg M, Richter M, Datchev A, et al. Glycolytic pyruvate regulates P-glycoprotein expression in multicellular tumor spheroids via modulation of the intracellular redox state. J Cell Biochem. 2010;109:434–446. doi: 10.1002/jcb.22422. [DOI] [PubMed] [Google Scholar]
- 81.Wagner W, Ciszewski WM, Kania KD. L- and D-lactate enhance DNA repair and modulate the resistance of cervical carcinoma cells to anticancer drugs via histone deacetylase inhibition and hydroxycarboxylic acid receptor 1 activation. Cell Commun Signal. 2015;13:36. doi: 10.1186/s12964-015-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 83.Vanhove K, Derveaux E, Graulus GJ, et al. Glutamine addiction and therapeutic strategies in lung cancer. Int J Mol Sci. 2019;20:252. doi: 10.3390/ijms20020252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wangpaichitr M, Wu C, Li YY, et al. Exploiting ROS and metabolic differences to kill cisplatin resistant lung cancer. Oncotarget. 2017;8:49275. doi: 10.18632/oncotarget.17568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eng CH, Yu K, Lucas J, White E, Abraham RT. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy. Sci Signal. 2010;3:ra31. doi: 10.1126/scisignal.2000911. [DOI] [PubMed] [Google Scholar]
- 86.Yang L, Achreja A, Yeung TL, et al. Targeting stromal glutamine synthetase in tumors disrupts tumor microenvironment-regulated cancer cell growth. Cell Metab. 2016;24:5. doi: 10.1016/j.cmet.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SY, Jeon HM, Ju MK, et al. Dlx-2 and glutaminase upregulate epithelial-mesenchymal transition and glycolytic switch. Oncotarget. 2016;7:7925–7939. doi: 10.18632/oncotarget.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mertz JA, Conery AR, Bryant BM, et al. Targeting Myc dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shimamura T, Chen Z, Soucheray M, et al. Efficacy of BET bromodomain inhibition in KRAS-mutant non-small cell lung cancer. Clin Cancer Res. 2013;19:6183–6192. doi: 10.1158/1078-0432.CCR-12-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lenhart R, Kirov S, Desilva H, et al. Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol Cancer Ther. 2015;14:2167–2174. doi: 10.1158/1535-7163. [DOI] [PubMed] [Google Scholar]
- 91.Riveiro ME, Astorgues-Xerri L, Vazquez R, et al. OTX015 (MK-8628), a novel BET inhibitor, exhibits antitumor activity in non-small cell and small cell lung cancer models harboring different oncogenic mutations. Oncotarget. 2016;7:84675–84687. doi: 10.18632/oncotarget.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fiorentino FP, Marchesi I, Schröder C, Schmidt R, Yokota J, Bagella L. BET-inhibitor I-BET762 and PARP-inhibitor talazoparib synergy in small cell lung cancer cells. Int J Mol Sci. 2020;21:9595. doi: 10.3390/IJMS21249595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Piha-Paul SA, Hann CL, French CA, et al. Phase 1 study of molibresib (GSK525762), a bromodomain and extra-terminal domain protein inhibitor, in NUT carcinoma and other solid tumors. JNCI Cancer Spectr. 2019;4:kz093. doi: 10.1093/jncics/pkz093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo J, Liu Y, Lv J, et al. BCL6 confers KRAS-mutant non–small-cell lung cancer resistance to BET inhibitors. J Clin Invest. 2021;131 doi: 10.1172/JCI133090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen D, Zhao Z, Huang Z, et al. Super enhancer inhibitors suppress Myc driven transcriptional amplification and tumor progression in osteosarcoma. Bone Res. 2018;6:11. doi: 10.1038/s41413-018-0009-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Christensen CL, Kwiatkowski N, Abraham BJ, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Padmanabhan J, Saha B, Powell C, Mo Q, Perez BA, Chellappan S. Inhibitors targeting CDK9 show high efficacy against osimertinib and AMG510 resistant lung adenocarcinoma cells. Cancers. 2021;13:3906. doi: 10.3390/cancers13153906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Calabrese DR, Chen X, Leon EC, et al. Chemical and structural studies provide a mechanistic basis for recognition of the Myc G-quadruplex. Nat Commun. 2018;9:4229. doi: 10.1038/s41467-018-06315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Local A, Zhang H, Benbatoul KD, et al. APTO-253 stabilizes G-quadruplex DNA, inhibits MYC expression, and induces DNA damage in acute myeloid leukemia cells. Mol Cancer Ther. 2018;17:1177–1186. doi: 10.1158/1535-7163.MCT-17-1209. [DOI] [PubMed] [Google Scholar]
- 100.Cercek A, Wheler J, Murray PE, Zhou S, Saltz L. Phase 1 study of APTO-253 HCl, an inducer of KLF4, in patients with advanced or metastatic solid tumors. Invest New Drugs. 2015;33:1086–1092. doi: 10.1007/s10637-015-0273-z. [DOI] [PubMed] [Google Scholar]
- 101.Pandya N, Kumar A. Piperine analogs arrest c-myc gene leading to downregulation of transcription for targeting cancer. Sci Rep. 2021;11:22909. doi: 10.1038/s41598-021-01529-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pawłowska M, Kulesza J, Augustin E. c-Myc protein level affected by unsymmetrical bisacridines influences apoptosis and senescence induced in HCT116 colorectal and H460 lung cancer cells. Int J Mol Sci. 2022;23:3061. doi: 10.3390/ijms23063061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang S, Awadasseid A, Narva S, et al. Anti-cancer activity of benzoxazinone derivatives via targeting c-Myc G-quadruplex structure. Life Sci. 2020;258 doi: 10.1016/J.LFS.2020.118252. [DOI] [PubMed] [Google Scholar]
- 104.Balaratnam S, Schneekloth JS. Transcriptional regulation of MYC through G-quadruplex structures. Annu Rep Med Chem. 2020;54:361–407. doi: 10.1016/bs.armc.2020.05.002. [DOI] [Google Scholar]
- 105.Wolfe AL, Singh K, Zhong Y, et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Davis SL, Leal AD, Messersmith WA, et al. A phase Ib study of the combination of alisertib (Aurora A kinase inhibitor) and MLN0128 (dual TORC1/2 inhibitor) in patients with advanced solid tumors, final expansion cohort data. 2022;40(16 Suppl):3112–3112. doi: 10.1200/JCO.2022.40.16_SUPPL.3112.
- 107.Pourdehnad M, Truitt ML, Siddiqi IN, Ducker GS, Shokat KM, Ruggero D. Myc and mTOR converge on a common node in protein synthesis control that confers synthetic lethality in Myc-driven cancers. Proc Natl Acad Sci USA. 2013;110:11988–11993. doi: 10.1073/pnas.1310230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Voss MH, Gordon MS, Mita M, et al. Phase 1 study of mTORC1/2 inhibitor sapanisertib (TAK-228) in advanced solid tumours, with an expansion phase in renal, endometrial or bladder cancer. Br J Cancer. 2020;123:1590–1598. doi: 10.1038/s41416-020-01041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hays J, Song Z, Paik P, et al. Results from the NCI-MATCH ECOG-ACRIN Trial (EAY131) – phase 2 study of MLN0128 (TAK-228) in patients with tumors with TSC1 or TSC2 mutations: sub-protocol EAY131-M. Eur J Cancer. 2022;174:S25. doi: 10.1016/S0959-8049(22)00870-X. [DOI] [Google Scholar]
- 110.Wiegering A, Uthe FW, Jamieson T, et al. Targeting translation initiation bypasses signaling crosstalk mechanisms that maintain high MYC levels in colorectal cancer. Cancer Discov. 2015;5:768–881. doi: 10.1158/2159-8290.CD-14-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hashimoto A, Handa H, Hata S, et al. Inhibition of mutant KRAS-driven overexpression of ARF6 and Myc by an eIF4A inhibitor drug improves the effects of anti-PD-1 immunotherapy for pancreatic cancer. Cell Commun Signal. 2021;19:54. doi: 10.1186/s12964-021-00733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Castell A, Larsson LG. Targeting MYC translation in colorectal cancer. Cancer Discov. 2015;5:701–703. doi: 10.1158/2159-8290.CD-15-0660. [DOI] [PubMed] [Google Scholar]
- 113.Shafat Z, Ahmed MM, Almajhdi FN, Hussain T, Parveen S, Ahmed A. Identification of the key miRNAs and genes associated with the regulation of non-small cell lung cancer: a network-based approach. Genes. 2022;13:1174. doi: 10.3390/genes13071174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rosenfeld YB, Krumbein M, Yeffet A, et al. VICKZ1 enhances tumor progression and metastasis in lung adenocarcinomas in mice. Oncogene. 2019;38:4169–4181. doi: 10.1038/s41388-019-0715-8. [DOI] [PubMed] [Google Scholar]
- 115.Wallis N, Oberman F, Shurrush K, et al. Small molecule inhibitor of Igf2bp1 represses Kras and a pro-oncogenic phenotype in cancer cells. RNA Biol. 2022;19:26–43. doi: 10.1080/15476286.2021.2010983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mahauad-Fernandez WD, Felsher DW. The myc and ras partnership in cancer: indistinguishable alliance or contextual relationship? Cancer Res. 2020;80:3799–3802. doi: 10.1158/0008-5472.CAN-20-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107lc. [DOI] [PubMed] [Google Scholar]
- 118.Soucek L, Whitfield JR, Sodir NM, et al. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 2013;27:504–513. doi: 10.1101/gad.205542.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Deng Q, Wu M, Deng J. USP36 promotes tumor growth of non-small cell lung cancer via increasing KHK-A expression by regulating c-MYC-hnRNPH1/H2 axis. Hum Cell. 2022;35:694–704. doi: 10.1007/s13577-022-00677-6. [DOI] [PubMed] [Google Scholar]
- 120.Pan J, Deng Q, Jiang C, et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34:3957–3967. doi: 10.1038/onc.2014.327. [DOI] [PubMed] [Google Scholar]
- 121.Zhang L, Xu B, Qiang Y, et al. Overexpression of deubiquitinating enzyme USP28 promoted non-small cell lung cancer growth. J Cell Mol Med. 2015;19:799–805. doi: 10.1111/jcmm.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Prieto-Garcia C, Hartmann O, Reissland M, et al. Maintaining protein stability of ΔNp63 via USP28 is required by squamous cancer cells. EMBO Mol Med. 2020;12:e11101. doi: 10.15252/emmm.201911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Z, Zhao T, Li Z, et al. Discovery of [1,2,3]triazolo[4,5-d]pyrimidine derivatives as highly potent, selective, and cellularly active USP28 inhibitors. Acta Pharm Sin B. 2020;10:1476–1491. doi: 10.1016/j.apsb.2019.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zeng Y, Li N, Liu W, Zeng M, Cheng J, Huang J. Analyses of expressions and prognostic values of Polo-like kinases in non-small cell lung cancer. J Cancer Res Clin Oncol. 2020;146:2447–2460. doi: 10.1007/s00432-020-03288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li H, Wang H, Sun Z, Guo Q, Shi H, Jia Y. The clinical and prognostic value of polo-like kinase 1 in lung squamous cell carcinoma patients: immunohistochemical analysis. Biosci Rep. 2017;37 doi: 10.1042/bsr20170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ellis PM, Leighl NB, Hirsh V, et al. A randomized, open-label phase II trial of volasertib as monotherapy and in combination with standard-dose pemetrexed compared with pemetrexed monotherapy in second-line treatment for non–small-cell lung cancer. Clin Lung Cancer. 2015;16:457–465. doi: 10.1016/J.cllc.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 127.Awad MM, Chu QS, Gandhi L, et al. An open-label, phase II study of the polo-like kinase-1 (Plk-1) inhibitor, BI 2536, in patients with relapsed small cell lung cancer (SCLC) Lung Cancer. 2017;104:126–130. doi: 10.1016/j.lungcan.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 128.Stratmann JA, Sebastian M. Polo-like kinase 1 inhibition in NSCLC: mechanism of action and emerging predictive biomarkers. Lung Cancer. 2019;10:67–80. doi: 10.2147/LCTT.S177618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Du R, Huang C, Liu K, Li X, Dong Z. Targeting AURKA in cancer: molecular mechanisms and opportunities for cancer therapy. Mol Cancer. 2021;20:15. doi: 10.1186/S12943-020-01305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Melichar B, Adenis A, Lockhart AC, et al. Safety and activity of alisertib, an investigational aurora kinase A inhibitor, in patients with breast cancer, small-cell lung cancer, non-small-cell lung cancer, head and neck squamous-cell carcinoma, and gastro-oesophageal adenocarcinoma: a five-arm phase 2 study. Lancet Oncol. 2015;16:395–405. doi: 10.1016/S1470-2045(15)70051-3. [DOI] [PubMed] [Google Scholar]
- 131.Blakely CM, Gubens MA, Allen GM, et al. Phase I study of the aurora kinase A inhibitor alisertib in combination with osimertinib in EGFR-mutant lung cancer. J Clin Oncol. 2021;39(15_suppl):9074. doi: 10.1200/JCO.2021.39.15_suppl.9074. –9074. [DOI] [Google Scholar]
- 132.Nader CP, Cidem A, Verrills NM, Ammit AJ. Protein phosphatase 2A (PP2A): a key phosphatase in the progression of chronic obstructive pulmonary disease (COPD) to lung cancer. Respir Res. 2019;20:1–18. doi: 10.1186/S12931-019-1192-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Saddoughi SA, Gencer S, Peterson YK, et al. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Mol Med. 2013;5:105–121. doi: 10.1002/EMMM.201201283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Farrell AS, Pelz C, Wang X, et al. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol. 2013;33:2930–2949. doi: 10.1128/MCB.01455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sears RC. The life cycle of c-Myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–1137. doi: 10.4161/CC.3.9.1145. [DOI] [PubMed] [Google Scholar]
- 136.Campaner E, Rustighi A, Zannini A, et al. A covalent PIN1 inhibitor selectively targets cancer cells by a dual mechanism of action. Nat Commun. 2017;8:15772. doi: 10.1038/ncomms15772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hart JR, Garner AL, Yu J, et al. Inhibitor of Myc identified in a Kröhnke pyridine library. Proc Natl Acad Sci USA. 2014;111:12556–12561. doi: 10.1073/pnas.1319488111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Castell A, Yan Q, Fawkner K, et al. A selective high affinity Myc-binding compound inhibits Myc:Max interaction and Myc-dependent tumor cell proliferation. Sci Rep. 2018;8:10064. doi: 10.1038/s41598-018-28107-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Han H, Jain AD, Truica MI, et al. Small-molecule Myc inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell. 2019;36:483–497. doi: 10.1016/j.ccell.2019.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; e15. 10.1016/j.ccell.2019.10.001.
- 140.Massó-Vallés D, Soucek L. Blocking Myc to treat cancer: reflecting on two decades of omomyc. Cells. 2020;9:883. doi: 10.3390/CELLS9040883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Beaulieu ME, Jauset T, Massó-Vallés D, et al. Intrinsic cell-penetrating activity propels omomyc from proof of concept to viable anti-myc therapy. Sci Transl Med. 2019;11:eaar5012. doi: 10.1126/scitranslmed.aar5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Omomyc as the first MYC-targeted therapy to successfully complete a phase I clinical trial – VHIO. 2023. Available from: https://vhio.net/2022/10/26/omomyc-as-the-first-myc-targeted-therapy-to-successfully-complete-a-phase-i-clinical-trial/. [Accessed on February 27, 2023].