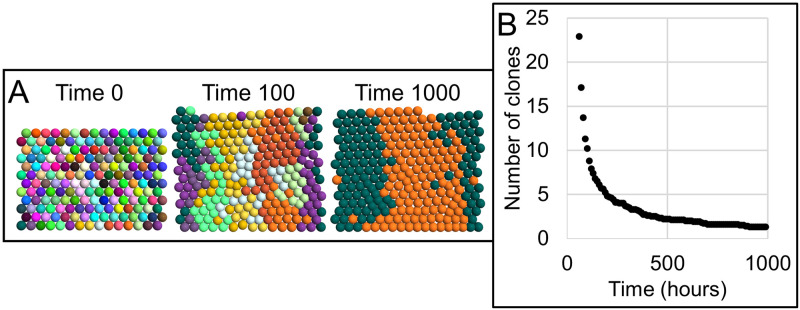
Fig 9. Two-dimensional agent-based model of cell proliferation and differential in the colonic crypt.
A: Cells represented as particles are arranged in a sheet as if fixed on an unfolded cylindrical surface, where the upper boundary is treated as the base of the crypt and periodic boundary conditions are applied along the horizontal direction, as in [16]. Each cell is assigned a state dynamics model of the cell cycle and unique clonal identification (visualized as a unique particle color). The cell cycle model of each cell progresses through G1, S, G2 and M phases with deterministic periods except for G1, the period of which is randomly selected from a normal distribution for each cell. When the cell cycle model of a cell transitions from the M phase to the G1 phase, the cell divides and copies its clonal identification to its progeny. Cells are removed when they reach the base of the crypt. B: Number of clones during simulation time, measured as the mean of ten simulation replicates. Over time, the crypt tends toward monoclonality.