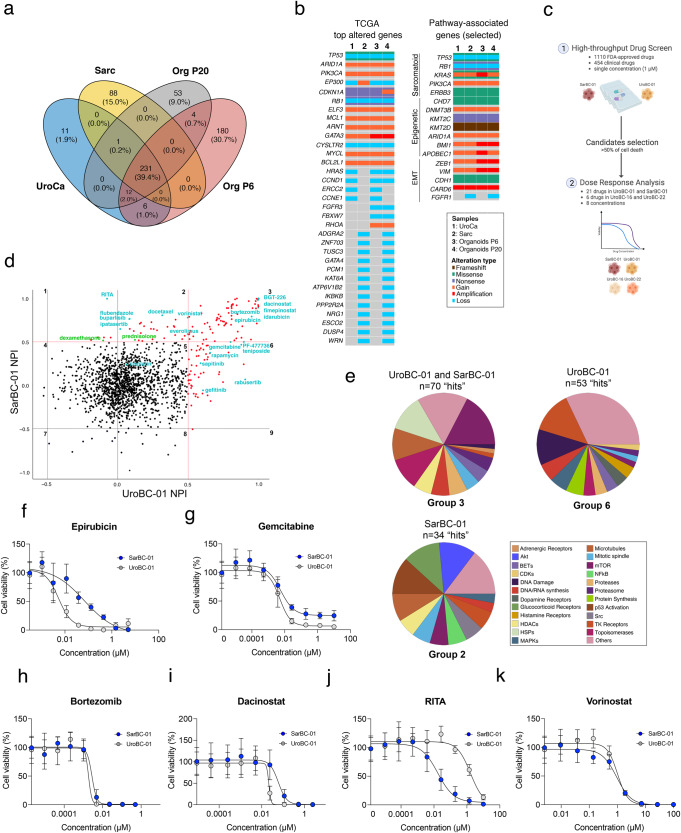
Fig. 3. Identification of genomic drivers and drug sensitivities in SarBC-01 organoids.
a Venn diagram depicting the number of shared mutations among SarBC-01 patient tumors’ components and derived organoids at early and late passages detected using whole exome sequencing (WES). Numbers in brackets indicate the proportion of shared mutations in each group. b Oncoplots depicting genomic alterations in the distinct SarBC-01 associated samples, as revealed by whole exome sequencing. Shown are alterations among the top 100 genes commonly mutated in BC (TCGA, left panel) and among selected genes associated with sarcomatoid cancers, epigenetic, and EMT pathways (right panel). Only alterations found in at least two samples are represented. A complete list of genomic alterations can be found in Supplementary Data 1. c Schematic representation of the workflow used for the high-throughput drug screening and subsequent dose response analyses. d Dot plot depicting normalized percentage inhibition (NPI) measured for SarBC-01 and UroBC-01, each dot representing one drug. “Hits” are defined as compounds associated with >50% inhibition as compared to negative controls (red dots, above the red line threshold). Compounds with inhibitory effects (“hits”) in UroBC-01 only (top right, group 6), SarBC-01 only (bottom left, group 2), and in both lines (top left, group 3) were identified. Drugs that have been further tested in dose response analyses are labelled in cyan. Labelled in green are the glucocorticoids dexamethasone and prednisolone. e Pie charts summarizing the mode of action categories of drugs identified as hits for both UroBC-01 and SarBC-01 (top left, group 3), UroBC-01 only (top right, group 6), and SarBC-01 only (bottom left, group 2). f–k Dose-responses curves for SarBC-01 and UroBC-01 organoids treated with a selected panel of drugs. Examples include standard of care compounds (f, g), drugs identified as “hit” in both SarBC-01 and UroBC-01 lines (h, i) or in SarBC-01 only (j, k). IC50 and GR50 values for all tested compounds are reported in Supplementary Data 3.