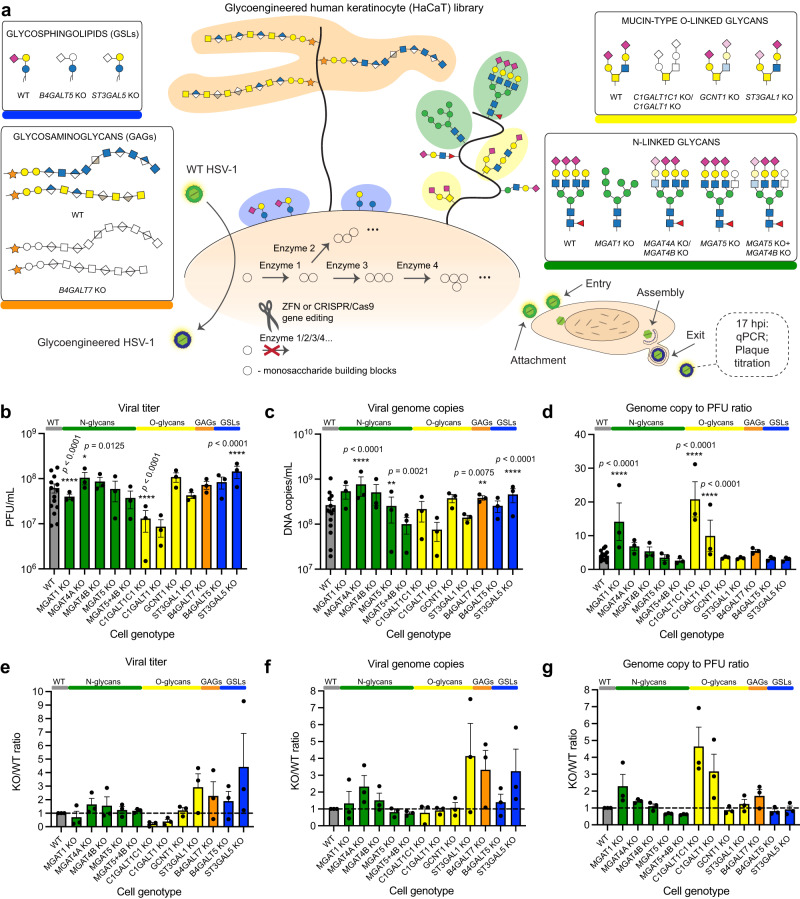
Fig. 1. Human keratinocyte library for probing host-pathogen interplay.
a Common cell surface glycoconjugates are depicted, with investigated classes of glycans highlighted in boxes. Structures are drawn according to the symbol nomenclature for glycans (SNFG). The glycoengineered cell library was generated by knocking out genes involved in respective biosynthetic pathways using CRISPR/Cas9 and ZFN technologies. b–d The array of cell lines was infected with MOI 10 of HSV-1 Syn17 + , and viral titers (b) and viral DNA content (c) in the media were measured at 17 hpi. Particle to PFU ratio is also shown (d). Data points show average values ± SEM from 3 independent experiments for each knock out (KO) cell line including paired WT data from a total of 14 experiments. Two-way ANOVA followed by Dunnett’s multiple comparison test was used to evaluate differences from WT (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). e–g Data from (b–d) is shown as WT-normalized mean + SEM of 3 independent experiments for each KO cell line. Source data are provided as a Source Data file for all graphs.