Abstract
The implementation of low-dose chest CT for lung screening presents a crucial opportunity to advance lung cancer care through early detection and interception. In addition, millions of pulmonary nodules are incidentally detected annually in the United States, increasing the opportunity for early lung cancer diagnosis. Yet, realization of the full potential of these opportunities is dependent on the ability to accurately analyze image data for purposes of nodule classification and early lung cancer characterization. This review presents an overview of traditional image analysis approaches in chest CT using semantic characterization as well as more recent advances in the technology and application of machine learning models using CT-derived radiomic features and deep learning architectures to characterize lung nodules and early cancers. Methodological challenges currently faced in translating these decision aids to clinical practice, as well as the technical obstacles of heterogeneous imaging parameters, optimal feature selection, choice of model, and the need for well-annotated image data sets for the purposes of training and validation, will be reviewed, with a view toward the ultimate incorporation of these potentially powerful decision aids into routine clinical practice.
© RSNA, 2023
See also the review “Milestones in CT: Past, Present, and Future” by McCollough and Rajiah in this issue.
Summary
The analysis of indeterminate pulmonary nodules by semantic annotation and, recently, quantitative approaches using machine learning and deep learning techniques are improving the distinction of malignant from benign nodules and may inform histologic and molecular features in early lung cancer.
Essentials
■ Advancements in chest CT have resulted in improvements in early lung cancer detection and the potential to reduce lung cancer mortality.
■ Owing to the increased use and capabilities of CT, millions of screen-detected and incidental pulmonary nodules are detected annually.
■ Semantic feature analysis, models developed with engineered radiomic features, and deep learning architectures present opportunities to improve our ability to determine which pulmonary nodules are malignant as well as their histologic and genetic makeup.
Introduction
Lung cancer is the leading cause of cancer-related death, accounting for more deaths than colon, breast, and prostate cancers combined (1). In 2011, the National Lung Screening Trial (NLST) demonstrated a 20% mortality benefit after three annual screenings with low-dose CT (LDCT) compared with chest radiography. This result showed that LDCT enables lung cancer detection at earlier, more treatable stages in people who smoke and are at high risk for lung cancer (2). Subsequent findings from the NELSON (Nederlands–Leuvens Longkanker Screenings Onderzoek) trial and a 10-year analysis of mortality in the Multicentric Italian Lung Detection trial have also confirmed lung cancer mortality benefits of 27%–39% in individuals screened with LDCT compared with no screening (3,4).
Despite the mortality benefits of LDCT screening in asymptomatic high-risk populations, a competing concern is the high prevalence of noncalcified indeterminate pulmonary nodules (IPNs), leading to high false-positive rates and potential negative consequences of downstream diagnostic testing. In the NLST, any noncalcified nodule with a maximum transverse diameter of 4 mm or larger was considered a positive screening result and mandated some form of follow-up beyond returning for an annual screen (2). Across all three screening rounds, screen positivity in the NLST LDCT arm was 24.2%, but was only 16.8% on the final screen because nodules observed to be stable over time could be categorized as negative (2).
Most nodules detected at lung screening were not cancer. In the NLST, 17 743 scans across three rounds contained a noncalcified nodule of maximum diameter of 4 mm or larger. In 64% of positive screens, the largest nodule was 7 mm or smaller, while this was the case for only 63 (10.5%) of 598 total cancers (100). Lung cancer rates increased with nodule size, most rapidly at nodule diameters of 10 mm or larger. Increasing the minimum size threshold for a positive screen to 5 mm maximum diameter would have resulted in a delayed or missed lung cancer diagnosis in 1.0% of cases while reducing the false-positive rate from 26.6% at baseline to 15.8%. Analyses by others using the NLST and other cohorts have confirmed reductions in false positivity rates using larger minimum size thresholds, but also lower test sensitivity (5,6).
The use of CT has increased substantially over time (as much as 53% over a 7-year period in one large health system [7]) and has propelled a rapid increase in the detection of IPNs. Although variously defined, IPNs are typically defined as focal opacities 6–30 mm in diameter without clearly benign patterns of calcification or intralesional fat, as in a hamartoma. Conservative estimates suggest that at least 1.5 million incidental pulmonary nodules are detected annually in the United States (7). Determining which IPNs are malignant among the vast majority that are benign is a critical unmet need that will only increase as the use of CT in routine diagnosis and screening becomes more common.
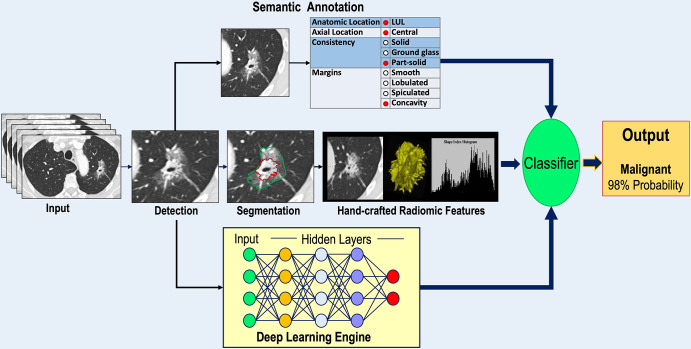
The morphologic features of nodules are known to influence the probability that nodules are lung cancer, and these features have been incorporated as variables into lung nodule diagnostic prediction models. Beyond semantic characterization of IPNs, advanced image analyses using engineered radiomic features, machine learning, and deep learning algorithms are increasingly being used to improve IPN classification as well as prognostication and optimal management of newly diagnosed lung cancers. Here, we present an overview of current image analysis approaches for refining indeterminate nodule classification and early lung cancer prognostication in newly diagnosed lung cancer (Fig 1).
Figure 1:
Diagram shows an overview of the major approaches to indeterminate pulmonary nodule classification and characterization of early lung cancer. A lung nodule detected on a CT scan may be annotated using semantic terms, undergo segmentation for extraction of radiomic features, or serve as input to a deep learning engine. The outputs of each analysis pathway are then submitted to some form of classifier to produce an output providing a probability of lung cancer or prediction of the histologic characteristics and/or genetic makeup of a known lung cancer. LUL = left upper lobe.
Semantic Feature Annotation
Semantic feature annotation is the labeling of an IPN by a human reader based on visual morphologic characteristics. Traditional features that may help differentiate benign and malignant IPNs include nodule-specific features such as size, border characteristics (smooth, spiculated, lobulated), and internal features (vacuoles, cavitation) and features of the perinodule surround (vascular convergence, pleural retraction, paracicatricial emphysema) (Fig 2). Visual assessment is the backbone of traditional image assessment, requires no special equipment, and continues to inform most current guidelines for nodule management. The Fleischner Society guidelines for the management of indeterminate nodules (8) and the Lung Imaging Reporting and Data System (Lung-RADS) guidelines developed by the American College of Radiology for screen-detected nodules (9) rely on semantic features to direct the timing and methods of follow-up of IPNs.
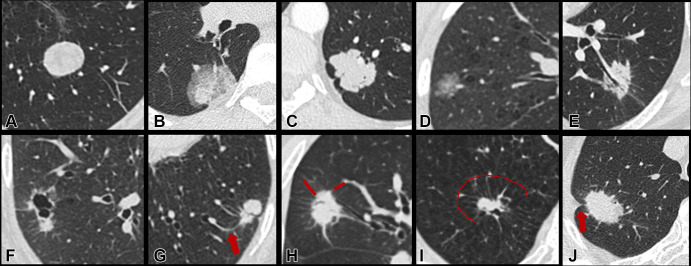
Figure 2:
Representative targeted CT images and corresponding semantic descriptors of lung nodules. A–F demonstrate different nodule consistencies and margin characteristics: (A) solid consistency, smooth margin; (B) part-solid consistency, smooth margin; (C) solid consistency, lobulated margin; (D) ground-glass consistency, lobulated margin; (E) solid consistency, spiculated margin; and (F) pericystic solid nodule, spiculated margin. G–J demonstrate different external characteristics of the surrounding perinodule lung parenchyma: (G) vascular convergence (arrow), (H) perinodule septal stretching (lines), (I) perinodule septal stretching with halo of paraseptal emphysema (line demarcates the emphysematous halo), and (J) pleural retraction (arrow).
Interobserver Agreement
Radiologists’ ability to identify and assess nodules is relatively robust when faced with variations in image acquisition and reconstruction parameters (10). While the software and hardware requirements for visual characterization of nodules are modest, human resource requirements in semantic annotation are high, and interobserver agreement on the designation of semantic features is variable. For example, concordance on the characterization of part-solid as opposed to pure ground-glass nodules has been reported as moderate across several studies, with Cohen κ values ranging from 0.51 to 0.67 (11). A radiologist’s ability to accurately assign semantic features to nodules improves with experience, and concordance between readers is highest among those with more years of post-training experience (12). The use of a standardized lexicon can also improve radiologist agreement in semantic annotation, as demonstrated with the adoption of the Lung-RADS reporting lexicon (13).
Role of Semantic Features in Existing Risk Models
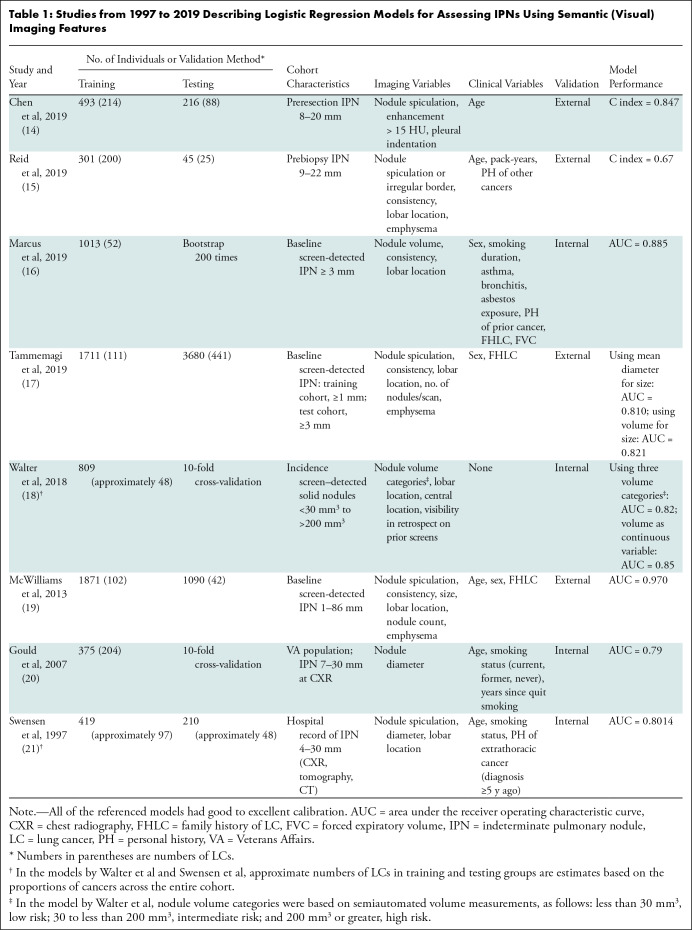
Semantic features factor in several logistic regression models of nodule malignancy (14–21). Table 1 provides a list of traditional models described in studies from 1997 to 2019 for assessing IPNs using semantic features. Earlier models by Gould et al (20) and Swensen et al (21) were based on nodules visible on chest radiographs, classic tomography images, or CT scans; more recent models were based on IPNs detected using CT. The models were developed in various clinical settings among populations with varying risks of lung cancer, including lung cancer screening populations, patients with IPNs identified through medical records reviews, or individuals undergoing surgical resection or nodule biopsy. With one exception (18), all of the models included imaging and clinical variables. Clinical variables differed across models, potentially because they were exclusionary criteria in some cohorts (eg, personal history of cancer), and in the case of smoking variables, their discriminatory ability was influenced by whether the cohorts under observation had explicit levels of smoking intensity or included individuals who had never smoked. Interestingly, chronic obstructive pulmonary disease, which is known to be associated with a three- to sixfold increase in lung cancer risk (22,23), was not a variable in most regression models.
Table 1:
Studies from 1997 to 2019 Describing Logistic Regression Models for Assessing IPNs Using Semantic (Visual) Imaging Features
Common imaging variables were nodule size, consistency, spiculation, location, number of nodules per scan, and enhancement following intravenous injection of contrast agent (Table 1). Nodule size is among the most discriminatory features in risk prediction models but was not included in three of the models (14,15,17). Tammemagi et al (17) developed two separate models using nodule mean diameter or volume. These were then tested on an NLST subsample in which benign nodules were matched to malignant nodules based on size, masking size as a distinguishing feature. Nodules in the models of Chen et al (14) and Reid et al (15) were of sufficiently high suspicion for cancer that they were all undergoing resection or biopsy, respectively. Nodules in these cohorts were within a narrower size range independent of histologic characteristics, obscuring the predictive association of size. Walter et al (18) explored benign and malignant new nodules seen on CT scans from incidence screening examinations and observed that morphologic characteristics that were associated with cancer in univariate analysis were not discriminatory in multivariate analysis. The authors found that size was more informative than other morphologic characteristics in new solid incidence nodules because size more closely reflects growth rate at incidence screening, as compared with prevalence nodules that may have been present for years prior to detection.
These models warrant a cautionary note when applying them to other IPN cohorts. Specifically, risk models perform optimally when applied to cases that are like the data sets in which they were developed. Selecting the appropriate model for a set of cases is critical to replicating model performance in real-world applications. For example, the Brock University model developed by McWilliams et al (19) was shown to retain high predictive performance in various screening cohorts that included small, largely benign nodules representing the spectrum of screen-detected lesions (Fig 3). However, when applied to a data set of predominantly large, high-risk nodules in which 69% were malignant, the area under the receiver operating characteristic curve (AUC) dropped to 0.693 (24). In clinical practice, prediction models are infrequently used in decision-making. A survey study among physicians in the American College of Chest Physicians reported a prediction model use rate of only 28% (25). Fortunately, physicians’ intuitive assessments of cancer risk perform well and have been shown to outperform prediction models in some instances (26,27).
Figure 3:
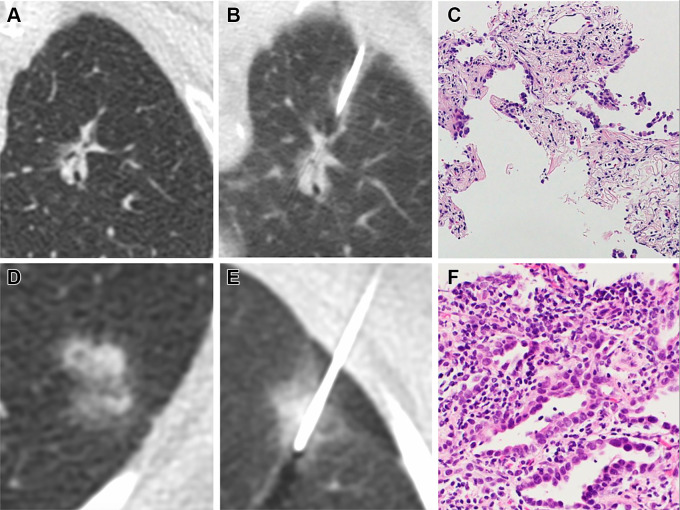
(A–C) Images in a 70-year-old woman without emphysema who is a current smoker. (A) Image from lung screening CT examination shows a part-solid nodule with a 26-mm mean diameter and a 9-mm solid component in the left upper lobe. The model developed by McWilliams et al (19) yielded a 78.1% probability of cancer given patient’s age, sex, and negative family history of lung cancer, and the nodule consistency, size, and location. (B) Image shows CT-guided percutaneous lung biopsy procedure. (C) Photomicrograph (hematoxylin-eosin stain) shows atypical epithelial cells with chronic inflammation and scarring. The patient underwent left upper lobectomy with resection of an invasive adenocarcinoma with a primarily acinar pattern, with papillary and lepidic components. (D–F) Images in a 68-year-old woman without significant emphysema who is a former smoker. (D) Image from low-dose CT examination of the chest shows part-solid nodule in the left lower lobe with a 14.1-mm mean diameter and a 13-mm solid component. The model developed by McWilliams et al (19) gave a 20.1% probability of malignancy. (E) Image shows CT-guided percutaneous lung biopsy procedure. (F) Photomicrograph (hematoxylin-eosin stain) shows well to moderately differentiated invasive lung adenocarcinoma with lepidic and focal acinar patterns.
Though routine in radiologic practice, semantic annotation is not quantitative. Moreover, the creation of large semantically annotated data sets is hampered by the fact that annotation is human resource intensive and does not scale well to the analysis of large, multi-institutional data sets. These limitations have led the imaging community to look to artificial intelligence to help analyze medical images.
Engineered Radiomic Features
Compared with the largely qualitative semantic descriptors of lung nodules, radiomic features are mathematically derived and provide a quantitative approach to nodule analysis. Several categories of mathematically derived features can be extracted from images: first-order (histogram-based), second-order, higher-order, transform-based, and shape features. This list is not comprehensive, as the number of extractable features has grown in the literature. Data mining and the use of radiomic features for classification tasks, prediction, and prognostication are of evolving interest in the lung cancer domain, primarily due to the fact that radiomics enables the noninvasive, objective assessment of the entire tumor at single or sequential time points, circumventing sampling biases inherent in histologic sampling.
The Radiomics Workflow
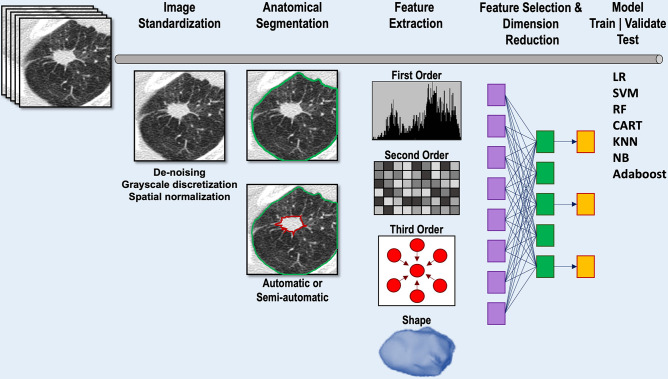
The prescribed mathematical formulae of engineered radiomic features are attractive in that they theoretically allow for features to be computed consistently between research laboratories. Several sequential steps are involved in radiomic analyses, including image acquisition, preprocessing to minimize image parameter heterogeneity, detection of the nodule of interest, nodule segmentation, extraction of radiomic features, feature selection and dimension reduction, and model fitting (Fig 4). In practice, the reproducibility of radiomic analysis is affected by each step in this pipeline. Here, we provide a high-level overview of the challenges associated with these steps as well as the potential uses of engineered radiomic features in nodule classification and lung cancer characterization.
Figure 4:
Diagram shows image analysis pipeline for conventional radiomic feature extraction and model development. Following input of the CT scan, image standardization may be performed to reduce image heterogeneity. Segmentation of the lung and nodule of interest can be performed automatically, semiautomatically, or manually. Following segmentation, radiomic features are extracted. Feature selection methods are applied to reduce the number of features while retaining as much feature variation as possible. The optimal radiomic features can then be input into one of a number of different machine learning models to train and validate the model. Model testing uses cross-validation, an internal subset of the data that has been set aside, or an external data set unrelated to the training data set. CART = classification and regression tree, KNN = k-nearest neighbor, LR = logistic regression, NB = naive Bayes, RF = random forest, SVM = support vector machine.
Image Acquisition and Reconstruction
The accuracy and reproducibility of radiomic feature analysis are inherently dependent on the choice of image acquisition and reconstruction parameters. Nonuniformity in CT imaging parameters, such as radiation dose, scanner pitch, section thickness, convolution kernel, and reconstruction algorithm, contributes to inconsistent, unstable radiomic features (28) and limits generalizability (29–32), and these issues are magnified in small and subsolid nodules (33,34). Radiomic shape features and first-order statistics appear to be more resilient to image parameter heterogeneity (28). There have been major efforts by the Image Biomarker Standardization Initiative, Quantitative Imaging Biomarkers Alliance, European Imaging Biomarkers Alliance, and others to establish standards for imaging biomarkers (35–37). However, uniform adoption is currently far from being realized, and image heterogeneity remains among the greatest obstacles to widescale dissemination of radiomics-based analyses.
Image Standardization, Harmonization, and Segmentation
To enhance the reproducibility of radiomic features, harmonization approaches can be applied directly to images or to the extracted radiomic features. Investigations have shown that harmonization techniques, by enhancing feature reproducibility, improve diagnostic and prediction models, although some techniques may adversely impact feature values and their discrimination ability (38). In the image domain, interpolation and resampling techniques are designed to produce image data of uniform spatial resolution since some radiomic features are dependent on voxel size (39). Similarly, denoising (40) and gray-scale discretization methods (41) are often applied to reconstructed images. Harmonization techniques can also be applied after radiomic features have been computed. For example, ComBat is an algorithm used to transform feature distributions that differ due to scanner and protocol use so that they are more similar (42,43).
Nodule segmentation approaches may be manual, semiautomatic, or fully automatic. Segmentation results can be highly variable, particularly for subsolid nodules or nodules with poorly defined margins (44), and may vary with the segmentation algorithm (45), which also adversely affects the reproducibility of some radiomic features. Manual segmentation can be highly labor intensive and does not scale for large studies; some experts advocate for radiologists to overread 5%–10% of such segmentations to enhance accuracy (46).
Feature Extraction, Selection, and Model Development
Open-source platforms such as the Quantitative Imaging Feature Pipeline (47), Computational Environment for Radiological Research (48), and PyRadiomics (49), among others, have been developed to extract a wide variety of engineered features using standardized definitions. Care should be taken when attempting to reproduce radiomic feature values across tools because, while efforts have improved the standardization of radiomic features, variations in values for some features remain due to implicit assumptions made about the input data or the ways in which certain mathematical operations or parameters are implemented by tool developers (50).
Given the large number of engineered radiomic features that can be extracted from a region of interest, some form of feature selection and dimension reduction is mandatory to minimize overfitting, which occurs when the model is too closely fit to the training data set and performs poorly when applied to other data sets. Both feature selection and dimension reduction seek to reduce the number of features by selecting the most highly relevant, nonredundant features, eliminating highly correlated or irrelevant features that contribute to noise.
Once extracted, radiomic features can be applied to models independently or in combination with additional clinical data such as demographics, histologic characteristics, or molecular tests to classify indeterminate nodules or predict the histologic characteristics, mutation status, or prognosis of known tumors. Several machine learning approaches have been used, including logistic regression, random forests, support vector machines, classification and regression trees, naive Bayes, k-nearest neighbor, and adaptive boosting, often grouped into families of linear, nonlinear, and ensemble classifiers.
Potential of Radiomic Models for Early Detection and Characterization
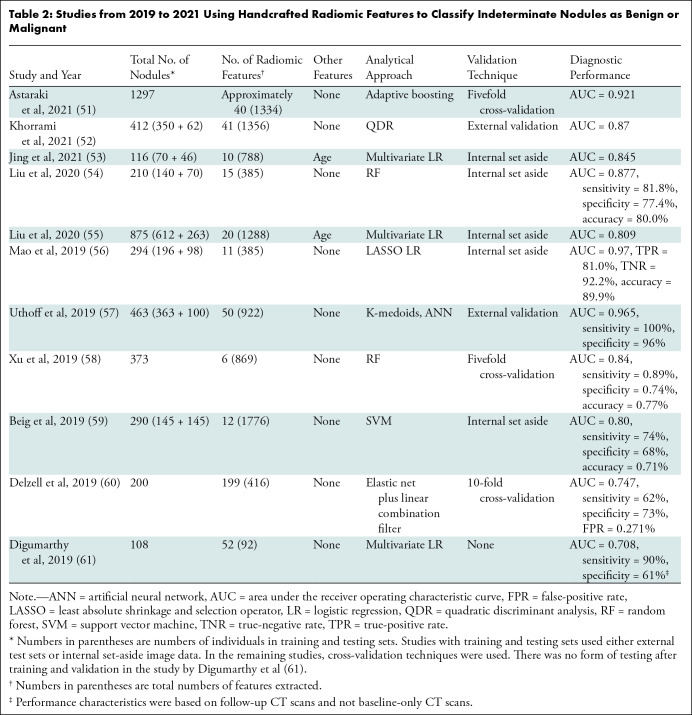
Table 2 summarizes recent studies on the performance of radiomic features in machine learning algorithms to distinguish benign and malignant IPNs (51–61). In these models, external data sets, which provide the most rigorous test of performance, were used in only two studies; in the remainder, test data sets were generated using resampling techniques, where different subsets of one data set are used to train and test the model. Both the feature selection method for dimension reduction and type of classifier used will affect model performance. Delzell et al (60) compared the performance of combinations of six feature selection methods and 12 classifiers in classifying 200 indeterminate nodules 5–30 mm in diameter. Their best-performing feature selection and classifier combination achieved an AUC of 0.747 (±0.111 [SD]); when the demographic variables age, sex, and pack-years (number of packs smoked per day times number of years smoked) were added to the model, the AUC increased to 0.854, underscoring the potential complementary nature of clinical and radiomic features (Table 2). The work by Delzell et al (60) highlighted additional points: (a) feature reduction prior to model training improved predictive performance; (b) different modeling approaches yielded different levels of performance, suggesting that in classification studies, different model types should be explored; and (c) the combined use of nodule and perinodule lung features improved discrimination. The incremental benefits of perinodule radiomic features have also been observed in other studies (57–59,62). Uthoff et al (57) described a pipeline in which various combinations of nodule, border, and perinodule radiomic features were used to train an ensemble of 10 artificial neural networks. The top-performing model used features of the nodule and perinodule surround. In training on 363 nodules, this model achieved complete separation of benign and malignant nodules. In two external test data sets of 100 cases and 73 cases, the AUCs were 0.965 and 0.924, respectively. The top five radiomic features included two features from the nodule surround and three from the nodule itself. Mao et al (56) tested a quantitative radiomic model with logistic regression that classified small, solid lung nodules (6–15 mm in diameter) on lung screening CT scans as malignant or benign, achieving an AUC of 0.97, compared with an AUC of 0.77 for Lung-RADS.
Table 2:
Studies from 2019 to 2021 Using Handcrafted Radiomic Features to Classify Indeterminate Nodules as Benign or Malignant
Beyond applying radiomics to classifying IPNs, investigators have developed radiomics-based machine learning algorithms to better distinguish lung adenocarcinoma from squamous cell carcinoma. Using preoperative CT scans from 105 patients with histologically confirmed adenocarcinoma or squamous cell carcinoma, Tang et al (63) trained and tested five common machine learning classifiers and an ensemble classifier. The best-performing ensemble model achieved an AUC of 0.78 in the test cohort using both tumoral and peritumoral radiomic features.
Differentiating precursor glandular lesions (atypical adenomatous hyperplasia and adenocarcinoma in situ) and adenocarcinomas (minimally invasive and invasive) in subsolid lesions has been the subject of several studies (64–68). Zhu et al (64) evaluated a cohort of 120 patients (129 lesions) with pathologically confirmed pure ground-glass nodules in an effort to distinguish precursor lesions from minimally invasive adenocarcinoma. After feature selection, 18 of 2107 radiomic features were selected for model construction. The classification performance (AUC) of a logistic regression model in the test set was 0.872 (95% CI: 0.756, 0.988). In another study involving 1018 pathologically confirmed ground-glass nodules, Zhu et al (65) trained a logistic regression model using tumoral and peritumoral radiomic features to derive a “rad-score” to distinguish precursor lesions from adenocarcinoma. After feature selection, nine tumoral and seven peritumoral radiomic features were selected from a total of 2446 total features. In a set-aside test cohort, the AUC of the rad-score was 0.828, outperforming a model using clinical and semantic features. A nomogram that combined semantic lobulation and the radiomic features performed best, with an AUC of 0.835 (65). The Computer-Aided Nodule Assessment and Risk Yield, or CANARY, classifier has been validated to predict survival in patients with lung adenocarcinoma (69,70) and epidermal growth factor receptor mutation status (71). Additionally, CANARY has been used as a tool to correlate biologic behaviors with radiomic features (72). Other investigations have also shown the potential of radiomics-based models to identify epidermal growth factor receptor and other mutations in lung cancer (73–76).
Deep Learning
Unlike radiomic analyses, deep learning algorithms do not necessarily depend on nodule segmentation and can determine which features to extract without human intervention through a process called representation learning (77). If provided with sufficient data, deep learning algorithms can exceed the performance of models using handcrafted features, picking up patterns that may be too subtle for humans to detect (46).
Current State
Deep learning is an active development area for automated lung nodule detection and characterization (78). The dominant type of neural network used to process medical imaging is the convolutional neural network (CNN). Common CNN-based architectures include ResNet (79), DenseNet (80), RetinaNet (81), U-Net (82), and Inception (83). More recently, vision transformers (or ViTs) have emerged as an alternative to CNNs where images are split into patches, projected into a feature space, and then processed using an encoder with a self-attention mechanism that permits the transformer model to learn long-range dependencies across different regions of an image. Vision transformers have shown promise in outperforming CNNs when trained with large data sets (84). A variety of strategies have been developed to mitigate challenges with obtaining annotated data, including data augmentation (generating new training examples based on existing data) (85) and self-supervised learning (pretraining the model on a related task where labels are easier to obtain) (86).
In grand challenges such as LUNA16 (87) and the Kaggle Data Science Bowl 2017, which provided annotated image data sets to enable comparisons between different nodule detection and classification algorithms, the algorithms with the best performance used deep learning–based approaches. However, most deep learning algorithms have not been shown to generalize to different cohorts with nodules of varying sizes and consistency (88). Only two of the algorithms from the Kaggle Data Science Bowl 2017 did not perform significantly worse than 11 radiologists at estimating the probability of lung cancer in an enriched set of 300 LDCT scans (89). Moreover, model performance in classifying IPNs has only been explicitly examined in a few studies (90–92).
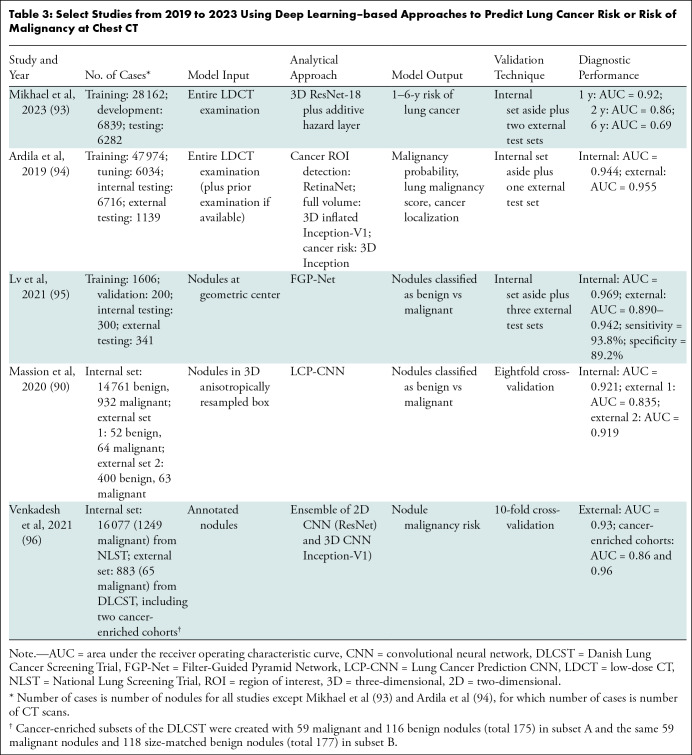
Estimation of lung cancer risk at the level of individual pulmonary nodules or entire volumetric CT examinations has emerged as a promising alternative to existing clinical risk models (Table 3). One emerging application of deep learning is to predict the risk of lung cancer in an individual based on scans from an entire volumetric LDCT examination. Most existing clinical risk models for estimating the likelihood of developing lung cancer use information on age, sex, exposures, smoking history, and family history. Mikhael et al (93) developed a deep learning–based model called Sybil that uses a ResNet-based encoder, an attention module, and an additive hazard layer to predict the risk of lung cancer 1–6 years after an LDCT screening examination. Data from 15 000 NLST CT-arm participants with 41 283 baseline and incidence LDCT examinations (of which 2080 examinations were positive and associated with cancer within 6 years) were used for model training and internal validation. The model was externally validated in two large institutional data sets. Sybil achieved an AUC of 0.92 for predicting 1-year lung cancer risk and 0.75 for predicting 6-year risk on the internal validation set and performed similarly on the external test sets. Sybil also achieved a lower false-positive rate than Lung-RADS (0.08 vs 0.10, P < .001, across all LDCT scans with visible nodules, n = 4201).
Table 3:
Select Studies from 2019 to 2023 Using Deep Learning–based Approaches to Predict Lung Cancer Risk or Risk of Malignancy at Chest CT
Ardila et al (94) developed a cancer detection algorithm that identified nodules on prior (when available) and current LDCT scans, processed up to two regions within scans containing a visible nodule, and predicted 1–2-year risk of lung cancer. Lv et al (95) developed a coupled human-machine classification approach for screen-detected and incidentally detected nodules in which most nodules were classified automatically by the algorithm and the remaining “ambiguous” nodules were classified with physician input. Venkadesh et al (96) developed and validated a model to distinguish malignant versus benign screen-detected nodules. The algorithm performed better than the existing PanCan nodule prediction model and trained radiologists. However, each of these studies used data from the NLST, which did not prospectively follow individual nodules to cancer outcomes. Also, studies often focus only on the AUC, omitting model calibration (the predicted probability of an event versus the observed probability), which is also an important performance measure. Massion et al (90) trained and internally validated a CNN based on the DenseNet architecture called the Lung Cancer Prediction CNN, or LCP-CNN, using 14 761 benign and 932 malignant solid and part-solid nodules that were 6 mm or larger from the NLST, and externally validated the algorithm in cohorts with IPNs, achieving higher discrimination performance than the Brock and Mayo risk prediction models. The LCP-CNN model was further evaluated in a multireader, multicase study that showed that sensitivity and specificity improved across all readers with the assistance of LCP-CNN (92).
Challenges and Opportunities
As with radiomics, differences in CT scanner manufacturer, radiation dose, convolution kernel, iterative reconstruction, and section thickness remain challenges to the diagnostic performance of deep learning algorithms. One study (97) compared the diagnostic performance of radiomics and deep learning algorithms in subtyping pulmonary adenocarcinoma amid changes in convolution kernel and strength of iterative reconstruction and found that deep learning was more susceptible to variability than radiomics.
The development of robust nodule detection and classification algorithms is facilitated by access to large data sets representative of the target population and with high-quality annotations. Many existing studies use large data sets such as those of the Lung Imaging Database Consortium or the NLST, but these data sets have limitations. The Lung Imaging Database Consortium data set contains a mixture of diagnostic CT and LDCT scans from patients who are not representative of a screening cohort. Only a small subset of nodules identified within the Lung Imaging Database Consortium cohort have pathologically confirmed diagnoses. On the other hand, the NLST represents a screening cohort with up to three annual screens and 7 years of clinical follow-up. However, suspicious nodules were neither uniquely tracked across scans nor explicitly tied to lung cancer diagnoses (ie, it is not always clear which nodule resulted in a given lung cancer diagnosis). The imaging community would greatly benefit from the availability of more data sets like the NELSON data set, where nodules were uniquely tracked and volumetric information was obtained, and from the sharing of expert-derived annotations (both region of interest and nodule boundary) from studies that have leveraged the NLST data set. Contributions of data from institutions with active lung cancer screening programs will also be critical to further train and validate models that can generalize across populations, given practice variability in the real world and the limitations of existing trials in representing the individuals who may benefit the most from screening.
Beyond Diagnosis
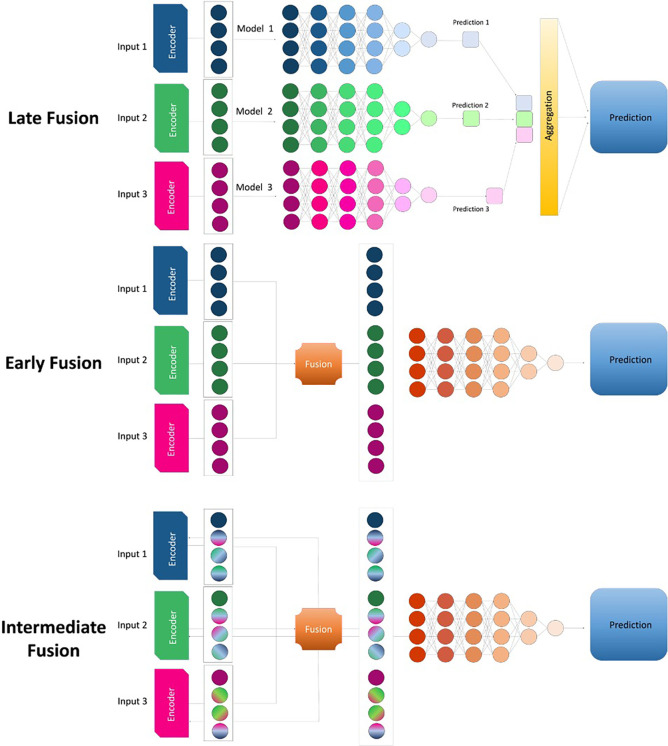
The role of medical imaging in lung cancer does not end with diagnosis. During the process of nodule characterization and management, data including clinical information, radiologic images, histopathologic characteristics, blood biomarker tests, and outcomes (eg, overall survival, recurrence-free survival) are generated. Imaging, either alone or combined with molecular and tissue-based biomarkers, can inform the appropriate next steps in treatment, help monitor recurrence, and inform outcomes. For example, Lian et al (98) showed that a vision transformer combined with a graph neural network could predict overall survival and recurrence-free survival in patients with early-stage lung cancer. With the increasing complexity of available data sets and computational capabilities, researchers are now investigating ways to effectively integrate different data types that span biologic scales (Fig 5). Various approaches, including late, early, and intermediate fusion, have been proposed for fusing different data types, resulting in a powerful approach to creating a more holistic model for prognostication in patients with lung cancer (99).
Figure 5:
Diagram shows three approaches to fusing data types in the creation of a model. In the late fusion approach, independent models are built for each input, and the predictions from each model are combined to generate a single outcome prediction. In the early fusion approach, a shared representation is created from the input features, ignoring what modality the input features originate from, before the features are fed through the model. In the intermediate fusion approach, the model takes advantage of input modality information by learning intermodality correlations before learning a joint representation.
Conclusion
Human semantic annotation currently dominates our approach to nodule classification and lung cancer characterization but is ultimately unsuited to analyses of large-scale image data sets. Analytical machine learning approaches are quantitative and objective and have the potential to help inform which nodules are malignant, personalize therapies based on characterization of cancer histologic characteristics and molecular signature, and inform prognosis. In hand with the promise of radiomics-based machine learning and deep learning approaches are complexities that we are only beginning to address. The heterogeneity of imaging acquisition and reconstruction substantially affects radiomic and deep learning features. Short of the convergence of the imaging community on standardized protocols in clinical practice, the challenge is to normalize images prior to feature extraction or to reduce the variations of extracted features, which may sometimes obscure discriminatory features. Given the sheer magnitude of extractable features, feature selection and dimension reduction techniques are critical for machine learning models to be confidently applied to new data sets; we do not yet have ways to determine the optimal feature selection approach or classification algorithm for a given decision task or data set. Ultimately, the imaging community can adopt these features and machine learning models only by showing their resilience to variation in acquisition protocols, their contributions to knowledge in specific clinical contexts, and their reproducibility across different patient populations. As well, among the imperatives for the imaging community is the provision of publicly available, high-quality, annotated clinical and image data sets that can be used for training and validation. Ultimately, as the biologic underpinnings of these classification algorithms become comprehensible and plausible to humans, computational algorithms can pave the way for more personalized approaches to diagnosis and treatment.
Disclosures of conflicts of interest: A.E.P. Grants or contracts from American College of Radiology, National Institutes of Health, DECAMP-1 PLUS, Edwards Life Sciences, National Cancer Institute (U2C CA271898), and National Institute for Biomedical Imaging and Bioengineering (R01 EB031993); consultant for MedQIA; honoraria from UCLA Global Health; leadership roles with the American College of Radiology and RSNA; and associate editor for Radiology: Imaging Cancer. M.N.K. Grants or contracts from National Cancer Institute; royalties or licenses from Meru Biotechnologies; consulting fees from Meru Biotechnologies and Biodesix; and support for attending meetings and/or travel from National Cancer Institute and Biodesix. F.M. Grants or contracts from National Cancer Institute (R01 CA253923). D.R.A. Grants or contracts from National Institutes of Health, American College of Radiology, DECAMP-1 PLUS, Kaiser Family Foundation, Watch the Spot Trial, National Cancer Institute (U2C CA271898), and National Institute for Biomedical Imaging and Bioengineering (R01 EB031993) and honoraria for presentations from Vanderbilt University. W.H. Grants or contracts from National Cancer Institute (U2C CA271898) and National Institute of Biomedical Imaging and Bioengineering (R01 EB031993) and deputy editor for Radiology: Artificial Intelligence.
Abbreviations:
- AUC
- area under the receiver operating characteristic curve
- CNN
- convolutional neural network
- IPN
- indeterminate pulmonary nodule
- LDCT
- low-dose CT
- Lung-RADS
- Lung Imaging Reporting and Data System
- NLST
- National Lung Screening Trial
References
- 1. Siegel RL , Miller KD , Jemal A . Cancer statistics, 2020 . CA Cancer J Clin 2020. ; 70 ( 1 ): 7 – 30 . [DOI] [PubMed] [Google Scholar]
- 2. National Lung Screening Trial Research Team ; Aberle DR , Adams AM , et al . Reduced lung-cancer mortality with low-dose computed tomographic screening . N Engl J Med 2011. ; 365 ( 5 ): 395 – 409 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Koning HJ , van der Aalst CM , de Jong PA , et al . Reduced lung-cancer mortality with volume CT screening in a randomized trial . N Engl J Med 2020. ; 382 ( 6 ): 503 – 513 . [DOI] [PubMed] [Google Scholar]
- 4. Pastorino U , Silva M , Sestini S , et al . Prolonged lung cancer screening reduced 10-year mortality in the MILD trial: new confirmation of lung cancer screening efficacy . Ann Oncol 2019. ; 30 ( 7 ): 1162 – 1169 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pinsky PF , Gierada DS , Black W , et al . Performance of Lung-RADS in the national lung screening trial: a retrospective assessment . Ann Intern Med 2015. ; 162 ( 7 ): 485 – 491 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yip R , Henschke CI , Yankelevitz DF , Smith JP . CT screening for lung cancer: alternative definitions of positive test result based on the national lung screening trial and international early lung cancer action program databases . Radiology 2014. ; 273 ( 2 ): 591 – 596 . [DOI] [PubMed] [Google Scholar]
- 7. Gould MK , Tang T , Liu ILA , et al . Recent trends in the identification of incidental pulmonary nodules . Am J Respir Crit Care Med 2015. ; 192 ( 10 ): 1208 – 1214 . [DOI] [PubMed] [Google Scholar]
- 8. MacMahon H , Naidich DP , Goo JM , et al . Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017 . Radiology 2017. ; 284 ( 1 ): 228 – 243 . [DOI] [PubMed] [Google Scholar]
- 9. Lung-RADS Version 1.1. American College of Radiology . https://www.acr.org/-/media/ACR/Files/RADS/Lung-RADS/LungRADSAssessmentCategoriesv1-1.pdf?la=en. Published 2019. Accessed March 23, 2023 .
- 10. Hassani C , Ronco A , Prosper AE , Dissanayake S , Cen SY , Lee C . Forward-projected model-based iterative reconstruction in screening low-dose chest CT: comparison with adaptive iterative dose reduction 3D . AJR Am J Roentgenol 2018. ; 211 ( 3 ): 548 – 556 . [DOI] [PubMed] [Google Scholar]
- 11. Revel MP , Mannes I , Benzakoun J , et al . Subsolid lung nodule classification: a CT criterion for improving interobserver agreement . Radiology 2018. ; 286 ( 1 ): 316 – 325 . [DOI] [PubMed] [Google Scholar]
- 12. Rampinelli C , Minotti M , Ancona E , et al . Inter-observer agreement on the morphology of screening-detected lung cancer: beyond pulmonary nodules and masses . Eur Radiol 2019. ; 29 ( 7 ): 3862 – 3870 . [DOI] [PubMed] [Google Scholar]
- 13. van Riel SJ , Jacobs C , Scholten ET , et al . Observer variability for Lung-RADS categorisation of lung cancer screening CTs: impact on patient management . Eur Radiol 2019. ; 29 ( 2 ): 924 – 931 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen XB , Yan RY , Zhao K , et al . Nomogram for the prediction of malignancy in small (8-20 mm) indeterminate solid solitary pulmonary nodules in Chinese populations . Cancer Manag Res 2019. ; 11 : 9439 – 9448 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reid M , Choi HK , Han X , et al . Development of a risk prediction model to estimate the probability of malignancy in pulmonary nodules being considered for biopsy . Chest 2019. ; 156 ( 2 ): 367 – 375 . [DOI] [PubMed] [Google Scholar]
- 16. Marcus MW , Duffy SW , Devaraj A , et al . Probability of cancer in lung nodules using sequential volumetric screening up to 12 months: the UKLS trial . Thorax 2019. ; 74 ( 8 ): 761 – 767 . [DOI] [PubMed] [Google Scholar]
- 17. Tammemagi M , Ritchie AJ , Atkar-Khattra S , et al . Predicting malignancy risk of screen-detected lung nodules-mean diameter or volume . J Thorac Oncol 2019. ; 14 ( 2 ): 203 – 211 . [DOI] [PubMed] [Google Scholar]
- 18. Walter JE , Heuvelmans MA , Bock GH , et al . Characteristics of new solid nodules detected in incidence screening rounds of low-dose CT lung cancer screening: the NELSON study . Thorax 2018. ; 73 ( 8 ): 741 – 747 . [DOI] [PubMed] [Google Scholar]
- 19. McWilliams A , Tammemagi MC , Mayo JR , et al . Probability of cancer in pulmonary nodules detected on first screening CT . N Engl J Med 2013. ; 369 ( 10 ): 910 – 919 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gould MK , Ananth L , Barnett PG ; Veterans Affairs SNAP Cooperative Study Group . A clinical model to estimate the pretest probability of lung cancer in patients with solitary pulmonary nodules . Chest 2007. ; 131 ( 2 ): 383 – 388 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Swensen SJ , Silverstein MD , Ilstrup DM , Schleck CD , Edell ES . The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules . Arch Intern Med 1997. ; 157 ( 8 ): 849 – 855 . [PubMed] [Google Scholar]
- 22. Young RP , Hopkins RJ , Christmas T , Black PN , Metcalf P , Gamble GD . COPD prevalence is increased in lung cancer, independent of age, sex and smoking history . Eur Respir J 2009. ; 34 ( 2 ): 380 – 386 . [DOI] [PubMed] [Google Scholar]
- 23. Mannino DM , Aguayo SM , Petty TL , Redd SC . Low lung function and incident lung cancer in the United States: data from the First National Health and Nutrition Examination Survey follow-up . Arch Intern Med 2003. ; 163 ( 12 ): 1475 – 1480 . [DOI] [PubMed] [Google Scholar]
- 24. Hammer MM , Nachiappan AC , Barbosa EJM Jr . Limited utility of pulmonary nodule risk calculators for managing large nodules . Curr Probl Diagn Radiol 2018. ; 47 ( 1 ): 23 – 27 . [DOI] [PubMed] [Google Scholar]
- 25. Tanner NT , Brasher PB , Jett J , Silvestri GA . Effect of a rule-in biomarker test on pulmonary nodule management: a survey of pulmonologists and thoracic surgeons . Clin Lung Cancer 2020. ; 21 ( 2 ): e89 – e98 . [DOI] [PubMed] [Google Scholar]
- 26. Tanner NT , Porter A , Gould MK , Li XJ , Vachani A , Silvestri GA . Physician assessment of pretest probability of malignancy and adherence with guidelines for pulmonary nodule evaluation . Chest 2017. ; 152 ( 2 ): 263 – 270 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gupta S , Jacobson FL , Kong CY , Hammer MM . Performance of lung nodule management algorithms for Lung-RADS category 4 lesions . Acad Radiol 2021. ; 28 ( 8 ): 1037 – 1042 . [DOI] [PubMed] [Google Scholar]
- 28. Reiazi R , Abbas E , Famiyeh P , et al . The impact of the variation of imaging parameters on the robustness of computed tomography radiomic features: a review . Comput Biol Med 2021. ; 133 : 104400 . [DOI] [PubMed] [Google Scholar]
- 29. Emaminejad N , Wahi-Anwar MW , Kim GHJ , Hsu W , Brown M , McNitt-Gray M . Reproducibility of lung nodule radiomic features: multivariable and univariable investigations that account for interactions between CT acquisition and reconstruction parameters . Med Phys 2021. ; 48 ( 6 ): 2906 – 2919 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Midya A , Chakraborty J , Gönen M , Do RKG , Simpson AL . Influence of CT acquisition and reconstruction parameters on radiomic feature reproducibility . J Med Imaging (Bellingham) 2018. ; 5 ( 1 ): 011020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Berenguer R , Pastor-Juan MDR , Canales-Vázquez J , et al . Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters . Radiology 2018. ; 288 ( 2 ): 407 – 415 . [DOI] [PubMed] [Google Scholar]
- 32. Kumar V , Gu Y , Basu S , et al . Radiomics: the process and the challenges . Magn Reson Imaging 2012. ; 30 ( 9 ): 1234 – 1248 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu Y , Lu L , Sun SH , et al . Effect of CT image acquisition parameters on diagnostic performance of radiomics in predicting malignancy of pulmonary nodules of different sizes . Eur Radiol 2022. ; 32 ( 3 ): 1517 – 1527 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gao Y , Hua M , Lv J , et al . Reproducibility of radiomic features of pulmonary nodules between low-dose CT and conventional-dose CT . Quant Imaging Med Surg 2022. ; 12 ( 4 ): 2368 – 2377 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zwanenburg A , Vallières M , Abdalah MA , et al . The Image Biomarker Standardization Initiative: standardized quantitative radiomics for high-throughput image-based phenotyping . Radiology 2020. ; 295 ( 2 ): 328 – 338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang EP , Wang XF , Choudhury KR , et al . Meta-analysis of the technical performance of an imaging procedure: guidelines and statistical methodology . Stat Methods Med Res 2015. ; 24 ( 1 ): 141 – 174 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. deSouza NM , Achten E , Alberich-Bayarri A , et al . Validated imaging biomarkers as decision-making tools in clinical trials and routine practice: current status and recommendations from the EIBALL* subcommittee of the European Society of Radiology (ESR) . Insights Imaging 2019. ; 10 ( 1 ): 87 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mali SA , Ibrahim A , Woodruff HC , et al . Making radiomics more reproducible across scanner and imaging protocol variations: a review of harmonization methods . J Pers Med 2021. ; 11 ( 9 ): 842 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shur JD , Doran SJ , Kumar S , et al . Radiomics in oncology: a practical guide . RadioGraphics 2021. ; 41 ( 6 ): 1717 – 1732 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen J , Bermejo I , Dekker A , Wee L . Generative models improve radiomics performance in different tasks and different datasets: an experimental study . Phys Med 2022. ; 98 : 11 – 17 . [DOI] [PubMed] [Google Scholar]
- 41. Haga A , Takahashi W , Aoki S , et al . Standardization of imaging features for radiomics analysis . J Med Invest 2019. ; 66 ( 1.2 ): 35 – 37 . [DOI] [PubMed] [Google Scholar]
- 42. Mahon RN , Ghita M , Hugo GD , Weiss E . ComBat harmonization for radiomic features in independent phantom and lung cancer patient computed tomography datasets . Phys Med Biol 2020. ; 65 ( 1 ): 015010 . [DOI] [PubMed] [Google Scholar]
- 43. Cabini RF , Brero F , Lancia A , et al . Preliminary report on harmonization of features extraction process using the ComBat tool in the multi-center “Blue Sky Radiomics” study on stage III unresectable NSCLC . Insights Imaging 2022. ; 13 ( 1 ): 38 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hassani C , Varghese BA , Nieva J , Duddalwar V . Radiomics in pulmonary lesion imaging . AJR Am J Roentgenol 2019. ; 212 ( 3 ): 497 – 504 . [DOI] [PubMed] [Google Scholar]
- 45. Shen W , Zhou M , Yang F , Yang C , Tian J . Multi-scale convolutional neural networks for lung nodule classification . Lect Notes Comput Sci 2015. ; 24 : 588 – 599 . [DOI] [PubMed] [Google Scholar]
- 46. Tunali I , Gillies RJ , Schabath MB . Application of radiomics and artificial intelligence for lung cancer precision medicine . Cold Spring Harb Perspect Med 2021. ; 11 ( 8 ): a039537 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mattonen SA , Gude D , Echegaray S , Bakr S , Rubin DL , Napel S . Quantitative imaging feature pipeline: a web-based tool for utilizing, sharing, and building image-processing pipelines . J Med Imaging (Bellingham) 2020. ; 7 ( 4 ): 042803 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Apte AP , Iyer A , Crispin-Ortuzar M , et al . Technical note: extension of CERR for computational radiomics: a comprehensive MATLAB platform for reproducible radiomics research . Med Phys 2018. ; 45 ( 8 ): 3713 – 3720 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Griethuysen JJM , Fedorov A , Parmar C , et al . Computational radiomics system to decode the radiographic phenotype . Cancer Res 2017. ; 77 ( 21 ): e104 – e107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McNitt-Gray M , Napel S , Jaggi A , et al . Standardization in quantitative imaging: a multicenter comparison of radiomic features from different software packages on digital reference objects and patient data sets . Tomography 2020. ; 6 ( 2 ): 118 – 128 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Astaraki M , Yang G , Zakko Y , Toma-Dasu I , Smedby Ö , Wang C . A comparative study of radiomics and deep-learning based methods for pulmonary nodule malignancy prediction in low dose CT images . Front Oncol 2021. ; 11 : 737368 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khorrami M , Bera K , Thawani R , et al . Distinguishing granulomas from adenocarcinomas by integrating stable and discriminating radiomic features on non-contrast computed tomography scans . Eur J Cancer 2021. ; 148 : 146 – 158 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jing R , Wang J , Li J , et al . A wavelet features derived radiomics nomogram for prediction of malignant and benign early-stage lung nodules . Sci Rep 2021. ; 11 ( 1 ): 22330 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu Y , Balagurunathan Y , Atwater T , et al . Radiological image traits predictive of cancer status in pulmonary nodules . Clin Cancer Res 2017. ; 23 ( 6 ): 1442 – 1449 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu A , Wang Z , Yang Y , et al . Preoperative diagnosis of malignant pulmonary nodules in lung cancer screening with a radiomics nomogram . Cancer Commun (Lond) 2020. ; 40 ( 1 ): 16 – 24 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mao L , Chen H , Liang M , et al . Quantitative radiomic model for predicting malignancy of small solid pulmonary nodules detected by low-dose CT screening . Quant Imaging Med Surg 2019. ; 9 ( 2 ): 263 – 272 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uthoff J , Stephens MJ , Newell JD Jr , et al . Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT . Med Phys 2019. ; 46 ( 7 ): 3207 – 3216 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu Y , Lu L , E LN , et al . Application of radiomics in predicting the malignancy of pulmonary nodules in different sizes . AJR Am J Roentgenol 2019. ; 213 ( 6 ): 1213 – 1220 . [DOI] [PubMed] [Google Scholar]
- 59. Beig N , Khorrami M , Alilou M , et al . Perinodular and intranodular radiomic features on lung CT images distinguish adenocarcinomas from granulomas . Radiology 2019. ; 290 ( 3 ): 783 – 792 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Delzell DAP , Magnuson S , Peter T , Smith M , Smith BJ . Machine learning and feature selection methods for disease classification with application to lung cancer screening image data . Front Oncol 2019. ; 9 : 1393 . [Published correction appears in Front Oncol 2020;10:866.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Digumarthy SR , Padole AM , Rastogi S , et al . Predicting malignant potential of subsolid nodules: can radiomics preempt longitudinal follow up CT? Cancer Imaging 2019. ; 19 ( 1 ): 36 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Masquelin AH , Alshaabi T , Cheney N , Estépar RSJ , Bates JHT , Kinsey CM . Perinodular parenchymal features improve indeterminate lung nodule classification . Acad Radiol 2023. ; 30 ( 6 ): 1073 – 1080 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tang X , Huang H , Du P , Wang L , Yin H , Xu X . Intratumoral and peritumoral CT-based radiomics strategy reveals distinct subtypes of non-small-cell lung cancer . J Cancer Res Clin Oncol 2022. ; 148 ( 9 ): 2247 – 2260 . [DOI] [PubMed] [Google Scholar]
- 64. Zhu YQ , Liu C , Mo Y , et al . Radiomics for differentiating minimally invasive adenocarcinoma from precursor lesions in pure ground-glass opacities on chest computed tomography . Br J Radiol 2022. ; 95 ( 1134 ): 20210768 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhu M , Yang Z , Wang M , et al . A computerized tomography-based radiomic model for assessing the invasiveness of lung adenocarcinoma manifesting as ground-glass opacity nodules . Respir Res 2022. ; 23 ( 1 ): 96 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jiang Y , Xiong Z , Zhao W , et al . Computed tomography radiomics-based distinction of invasive adenocarcinoma from minimally invasive adenocarcinoma manifesting as pure ground-glass nodules with bubble-like signs . Gen Thorac Cardiovasc Surg 2022. ; 70 ( 10 ): 880 – 890 . [DOI] [PubMed] [Google Scholar]
- 67. Chen X , Yao B , Li J , Liang C , Qi R , Yu J . Feasibility of using high-resolution computed tomography features for invasiveness differentiation of malignant nodules manifesting as ground-glass nodules . Can Respir J 2022. ; 2022 : 2671772 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lv Y , Ye J , Yin YL , Ling J , Pan XP . A comparative study for the evaluation of CT-based conventional, radiomic, combined conventional and radiomic, and delta-radiomic features, and the prediction of the invasiveness of lung adenocarcinoma manifesting as ground-glass nodules . Clin Radiol 2022. ; 77 ( 10 ): e741 – e748 . [DOI] [PubMed] [Google Scholar]
- 69. Maldonado F , Boland JM , Raghunath S , et al . Noninvasive characterization of the histopathologic features of pulmonary nodules of the lung adenocarcinoma spectrum using computer-aided nodule assessment and risk yield (CANARY)—a pilot study . J Thorac Oncol 2013. ; 8 ( 4 ): 452 – 460 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Clay R , Rajagopalan S , Karwoski R , Maldonado F , Peikert T , Bartholmai B . Computer Aided Nodule Analysis and Risk Yield (CANARY) characterization of adenocarcinoma: radiologic biopsy, risk stratification and future directions . Transl Lung Cancer Res 2018. ; 7 ( 3 ): 313 – 326 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Clay R , Kipp BR , Jenkins S , et al . Computer-Aided Nodule Assessment and Risk Yield (CANARY) may facilitate non-invasive prediction of EGFR mutation status in lung adenocarcinomas . Sci Rep 2017. ; 7 ( 1 ): 17620 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Senosain MF , Zou Y , Novitskaya T , et al . HLA-DR cancer cells expression correlates with T cell infiltration and is enriched in lung adenocarcinoma with indolent behavior . Sci Rep 2021. ; 11 ( 1 ): 14424 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yamazaki M , Yagi T , Tominaga M , Minato K , Ishikawa H . Role of intratumoral and peritumoral CT radiomics for the prediction of EGFR gene mutation in primary lung cancer . Br J Radiol 2022. ; 95 ( 1140 ): 20220374 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Li S , Li Y , Zhao M , Wang P , Xin J . Combination of 18F-fluorodeoxyglucose PET/CT radiomics and clinical features for predicting epidermal growth factor receptor mutations in lung adenocarcinoma . Korean J Radiol 2022. ; 23 ( 9 ): 921 – 930 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Perez-Johnston R , Araujo-Filho JA , Connolly JG , et al . CT-based radiogenomic analysis of clinical stage I lung adenocarcinoma with histopathologic features and oncologic outcomes . Radiology 2022. ; 303 ( 3 ): 664 – 672 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Le NQK , Kha QH , Nguyen VH , Chen YC , Cheng SJ , Chen CY . machine learning-based radiomics signatures for EGFR and KRAS mutations prediction in non-small-cell lung cancer . Int J Mol Sci 2021. ; 22 ( 17 ): 9254 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hosny A , Parmar C , Quackenbush J , Schwartz LH , Aerts HJWL . Artificial intelligence in radiology . Nat Rev Cancer 2018. ; 18 ( 8 ): 500 – 510 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gu Y , Chi J , Liu J , et al . A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning . Comput Biol Med 2021. ; 137 : 104806 . [DOI] [PubMed] [Google Scholar]
- 79. He K , Zhang X , Ren S , Sun J . Deep residual learning for image recognition . In: Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) . Institute of Electrical and Electronics Engineers; ; 2016. : 770 – 778 . [Google Scholar]
- 80. Huang G , Liu Z , Maaten LVD , Weinberger KQ . Densely connected convolutional networks . In: Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) . Institute of Electrical and Electronics Engineers; ; 2017. : 2261 – 2269 . [Google Scholar]
- 81. Lin TY , Goyal P , Girshick R , He K , Dollar P . Focal loss for dense object detection . IEEE Trans Pattern Anal Mach Intell 2020. ; 42 ( 2 ): 318 – 327 . [DOI] [PubMed] [Google Scholar]
- 82. Ronneberger O , Fischer P , Brox T , eds. U-Net: convolutional networks for biomedical image segmentation . In: Navab N , Hornegger J , Wells W , Frangi A , eds. Medical image computing and computer-assisted intervention–MICCAI 2015. Lecture notes in computer science; vol 9351 . Cham, Switzerland: : Springer International Publishing; , 2015. . [Google Scholar]
- 83. Szegedy C , Liu W , Jia Y , et al . Going deeper with convolutions . In: Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) . Institute of Electrical and Electronics Engineers; ; 2015. : 1 – 9 . [Google Scholar]
- 84. Chen X , Wang X , Zhang K , et al . Recent advances and clinical applications of deep learning in medical image analysis . Med Image Anal 2022. ; 79 : 102444 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chlap P , Min H , Vandenberg N , Dowling J , Holloway L , Haworth A . A review of medical image data augmentation techniques for deep learning applications . J Med Imaging Radiat Oncol 2021. ; 65 ( 5 ): 545 – 563 . [DOI] [PubMed] [Google Scholar]
- 86. Krishnan R , Rajpurkar P , Topol EJ . Self-supervised learning in medicine and healthcare . Nat Biomed Eng 2022. ; 6 ( 12 ): 1346 – 1352 . [DOI] [PubMed] [Google Scholar]
- 87. Setio AAA , Traverso A , de Bel T , et al . Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge . Med Image Anal 2017. ; 42 : 1 – 13 . [DOI] [PubMed] [Google Scholar]
- 88. Min Y , Hu L , Wei L , Nie S . Computer-aided detection of pulmonary nodules based on convolutional neural networks: a review . Phys Med Biol 2022. ; 67 ( 6 ): 06TR01 . [DOI] [PubMed] [Google Scholar]
- 89. Jacobs C , Setio AAA , Scholten ET , et al . Deep learning for lung cancer detection on screening CT scans: results of a large-scale public competition and an observer study with 11 radiologists . Radiol Artif Intell 2021. ; 3 ( 6 ): e210027 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Massion PP , Antic S , Ather S , et al . Assessing the accuracy of a deep learning method to risk stratify indeterminate pulmonary nodules . Am J Respir Crit Care Med 2020. ; 202 ( 2 ): 241 – 249 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gao R , Li T , Tang Y , et al . Reducing uncertainty in cancer risk estimation for patients with indeterminate pulmonary nodules using an integrated deep learning model . Comput Biol Med 2022. ; 150 : 106113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kim RY , Oke JL , Pickup LC , et al . Artificial intelligence tool for assessment of indeterminate pulmonary nodules detected with CT . Radiology 2022. ; 304 ( 3 ): 683 – 691 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mikhael PG , Wohlwend J , Yala A , et al . Sybil: a validated deep learning model to predict future lung cancer risk from a single low-dose chest computed tomography . J Clin Oncol 2023. ; 41 ( 12 ): 2191 – 2200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ardila D , Kiraly AP , Bharadwaj S , et al . End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography . Nat Med 2019. ; 25 ( 6 ): 954 – 961 . [Published correction appears in Nat Med 2019;25(8):1319.] [DOI] [PubMed] [Google Scholar]
- 95. Lv W , Wang Y , Zhou C , et al . Development and validation of a clinically applicable deep learning strategy (HONORS) for pulmonary nodule classification at CT: a retrospective multicentre study . Lung Cancer 2021. ; 155 : 78 – 86 . [DOI] [PubMed] [Google Scholar]
- 96. Venkadesh KV , Setio AAA , Schreuder A , et al . Deep learning for malignancy risk estimation of pulmonary nodules detected at low-dose screening CT . Radiology 2021. ; 300 ( 2 ): 438 – 447 . [DOI] [PubMed] [Google Scholar]
- 97. Zhao W , Zhang W , Sun Y , et al . Convolution kernel and iterative reconstruction affect the diagnostic performance of radiomics and deep learning in lung adenocarcinoma pathological subtypes . Thorac Cancer 2019. ; 10 ( 10 ): 1893 – 1903 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lian J , Deng J , Hui ES , et al . Early stage NSCLS patients’ prognostic prediction with multi-information using transformer and graph neural network model . eLife 2022. ; 11 : e80547 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lipkova J , Chen RJ , Chen B , et al . Artificial intelligence for multimodal data integration in oncology . Cancer Cell 2022. ; 40 ( 10 ): 1095 – 1110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Gierada DS , Pinsky P , Nath H , Chiles C , Duan F , Aberle DR . Projected outcomes using different nodule sizes to define a positive CT lung cancer screening examination . J Natl Cancer Inst 2014. ; 106 ( 11 ): dju284 . [DOI] [PMC free article] [PubMed] [Google Scholar]