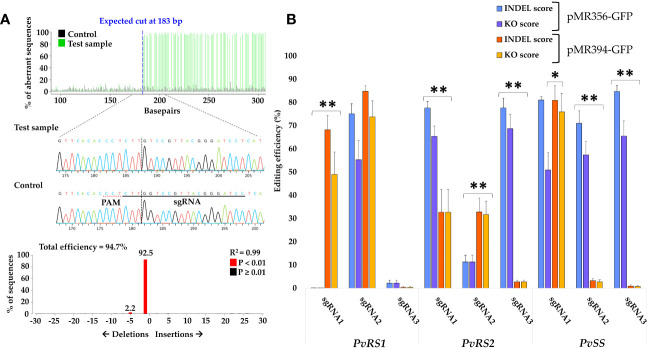
Figure 5.
Detection of mutations and summary of editing efficiency of 9 different sgRNAs within pMR356-GFP and pMR394-GFP targeting the RFO biosynthetic genes in P. vulgaris. (A) Overview of TIDE results, including observed mutation types, of one hairy root edited by sgRNA3 (pMR356-GFP) targeting Raffinose synthase 2 (PvRS2). (B) Summary of the mean sgRNA efficiencies in inducing insertions and deletions (INDEL score) and knockout mutations (KO score) in Raffinose synthase 1 (PvRS1), PvRS2 and Stachyose synthase (PvSS). The mean INDEL and KO scores for each sgRNA of pMR356-GFP and pMR394-GFP are shown. Error bars represent the standard error of the mean (n ≥ 15). A single asterisk (*) indicates that only the KO scores were significantly different (p < 0.05). Double asterisk (**) indicates that both the INDEL scores as well as the KO scores were significantly different (p < 0.01) based on a two-sample t-test assuming unequal variances. All gene editing analyses were performed on hairy roots derived from Method 3.