Abstract
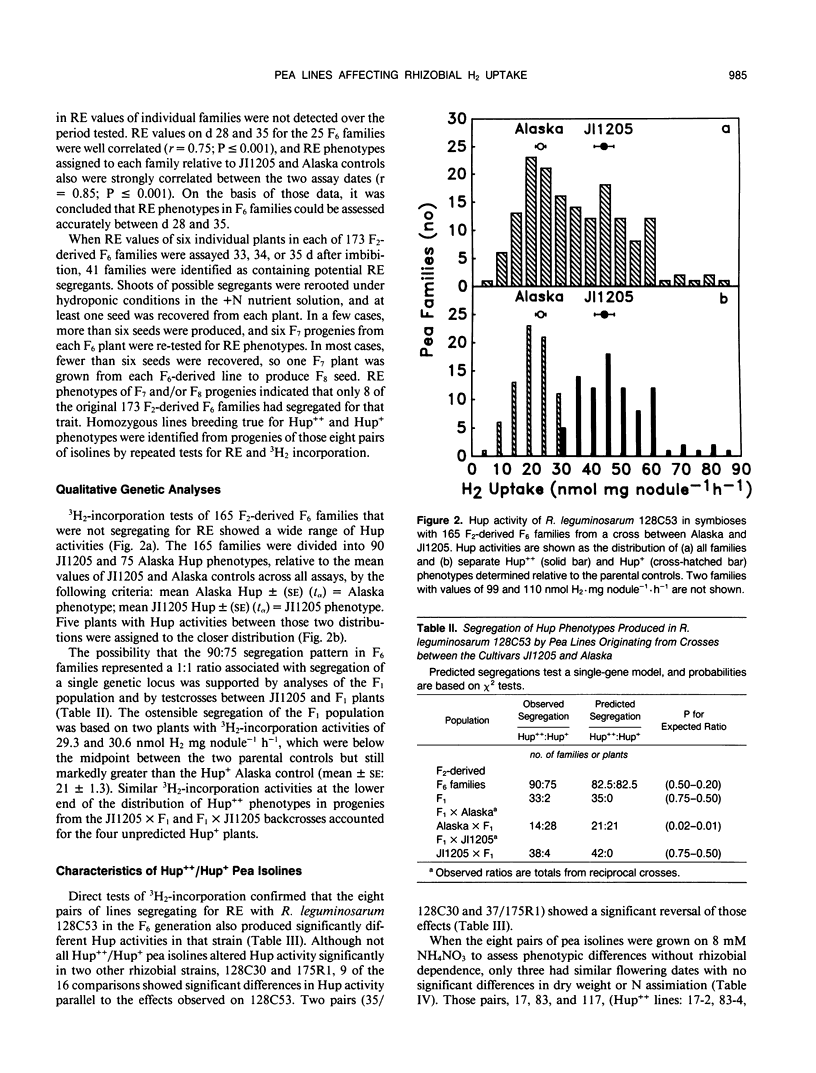
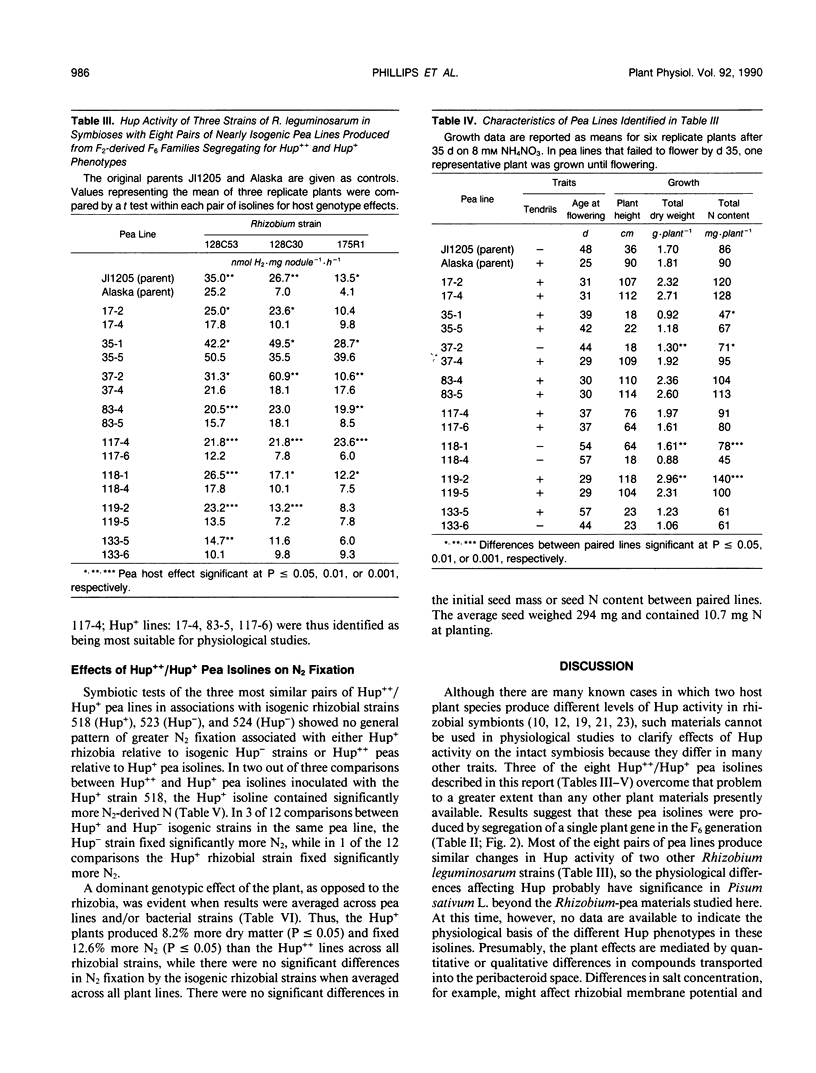
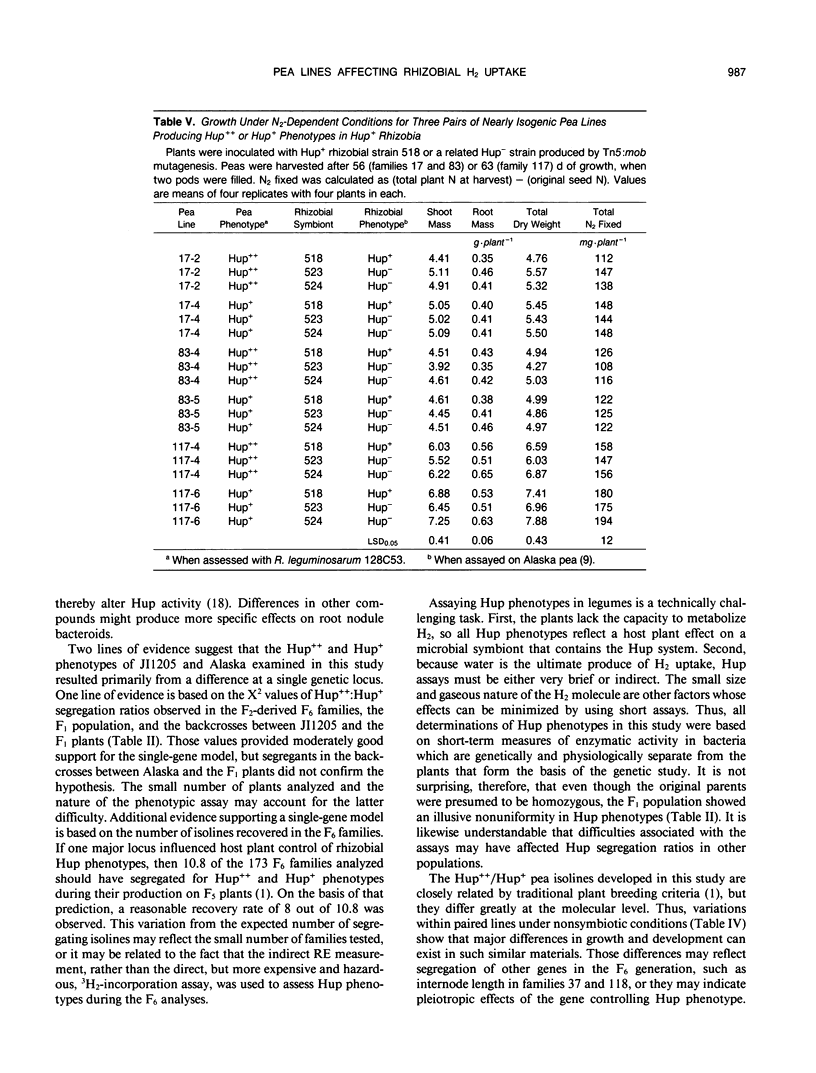
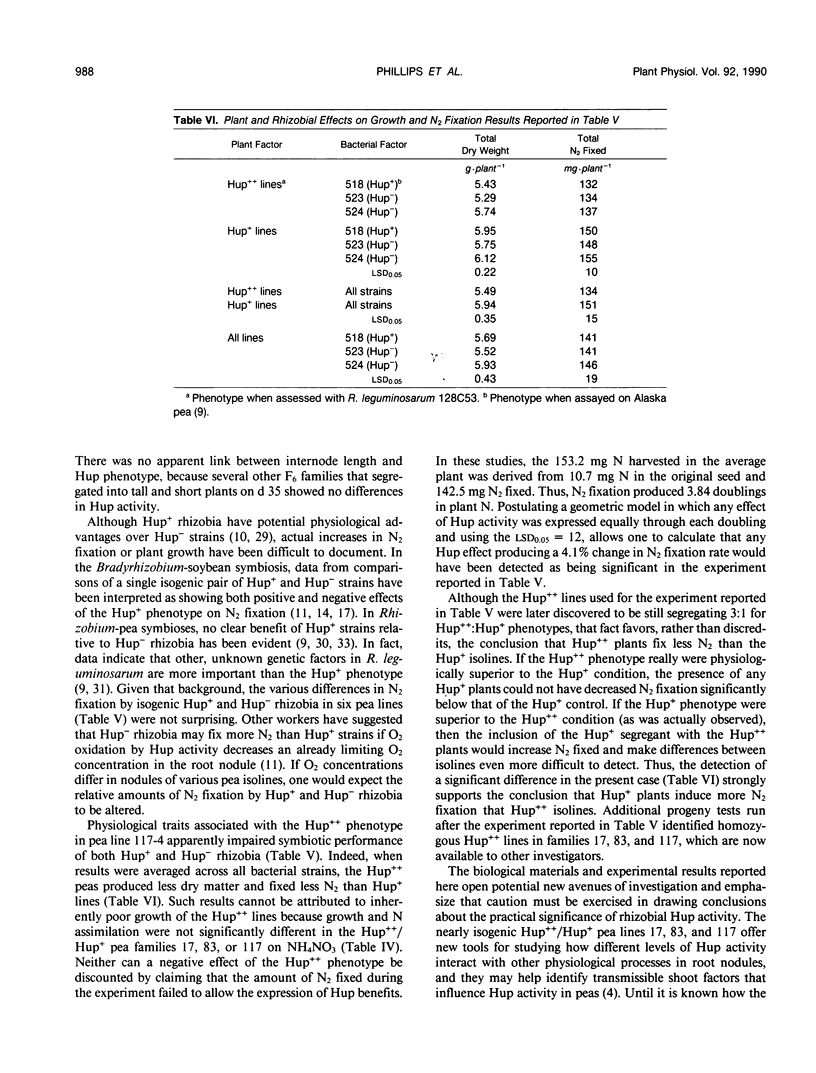
Some Rhizobium bacteria have H2-uptake (Hup) systems that oxidize H2 evolved from nitrogenase in leguminous root nodules. Pea (Pisum sativum L.) cultivars `JI1205' and `Alaska' produce high Hup (Hup++) and moderate Hup (Hup+) phenotypes, respectively, in Rhizobium leguminosarum 128C53. The physiological significance and biochemical basis of this host plant genetic effect are unknown. The purpose of this investigation was to advance basic Hup studies by developing nearly isogenic lines of peas that alter Hup phenotypes in R. leguminosarum strains containing hup genes. Eight pairs of nearly isogenic pea lines that produce Hup++ and Hup+ phenotypes in R. leguminosarum 128C53 were identified in 173 F2-derived F6 families produced from crosses between JI1205 and Alaska. Tests with the pea isolines and three strains of hup-containing R. leguminosarum showed that the isolines altered Hup activity significantly (P ≤ 0.05) in 19 of 24 symbiotic combinations. Analyses of Hup phenotypes in F6 families, the F1 population, and two backcrosses suggested involvement of a single genetic locus. Three of the eight pairs of isolines were identified as being suitable for physiological studies, because the two lines in each pair showed similar growth, N assimilation, and flowering traits under nonsymbiotic conditions. Tests of those lines under N2-dependent conditions with isogenic Hup+ and negligible Hup (Hup−) mutants of R. leguminosarum 128C53 showed that, in symbioses with Hup+ rhizobia, two out of three Hup++ pea lines decreased N2 fixation relative to Hup+ peas. In one of those cases, however, the Hup++ plant line also decreased fixation by Hup− rhizobia. When results were averaged across all rhizobia tested, Hup+ pea isolines had 8.2% higher dry weight (P ≤ 0.05) and fixed 12.6% more N2 (P ≤ 0.05) than Hup++ isolines. Pea lines described here may help identify host plant factors that influence rhizobial Hup activity and should assist in clarifying how Hup systems influence other physiological processes.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arp D. J. Rhizobium japonicum hydrogenase: purification to homogeneity from soybean nodules, and molecular characterization. Arch Biochem Biophys. 1985 Mar;237(2):504–512. doi: 10.1016/0003-9861(85)90303-0. [DOI] [PubMed] [Google Scholar]
- Bedmar E. J., Edie S. A., Phillips D. A. Host Plant Cultivar Effects on Hydrogen Evolution by Rhizobium leguminosarum. Plant Physiol. 1983 Aug;72(4):1011–1015. doi: 10.1104/pp.72.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedmar E. J., Phillips D. A. A transmissible plant shoot factor promotes uptake hydrogenase activity in Rhizobium symbionts. Plant Physiol. 1984 Jul;75(3):629–633. doi: 10.1104/pp.75.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethlenfalvay G. J., Phillips D. A. Variation in nitrogenase and hydrogenase activity of alaska pea root nodules. Plant Physiol. 1979 May;63(5):816–820. doi: 10.1104/pp.63.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulen W. A., LeComte J. R. The nitrogenase system from Azotobacter: two-enzyme requirement for N2 reduction, ATP-dependent H2 evolution, and ATP hydrolysis. Proc Natl Acad Sci U S A. 1966 Sep;56(3):979–986. doi: 10.1073/pnas.56.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham S. D., Kapulnik Y., Brewin N. J., Phillips D. A. Uptake Hydrogenase Activity Determined by Plasmid pRL6JI in Rhizobium leguminosarum Does Not Increase Symbiotic Nitrogen Fixation. Appl Environ Microbiol. 1985 Oct;50(4):791–794. doi: 10.1128/aem.50.4.791-794.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. O. Hydrogenase in legume root nodule bacteroids: occurrence and properties. Arch Mikrobiol. 1972;85(3):193–201. doi: 10.1007/BF00408844. [DOI] [PubMed] [Google Scholar]
- Drevon J. J., Kalia V. C., Heckmann M. O., Salsac L. Influence of the Bradyrhizobium japonicum Hydrogenase on the Growth of Glycine and Vigna Species. Appl Environ Microbiol. 1987 Mar;53(3):610–612. doi: 10.1128/aem.53.3.610-612.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisbrenner G., Evans H. J. Spectral evidence for a component involved in hydrogen metabolism of soybean nodule bacteroids. Plant Physiol. 1982 Dec;70(6):1667–1672. doi: 10.1104/pp.70.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal S. S., Rains D. W., Huffaker R. C. Determination of ammonium ion by fluorometry or spectrophotometry after on-line derivatization with o-phthalaldehyde. Anal Chem. 1988 Jan 15;60(2):175–179. doi: 10.1021/ac00153a016. [DOI] [PubMed] [Google Scholar]
- Haugland R. A., Cantrell M. A., Beaty J. S., Hanus F. J., Russell S. A., Evans H. J. Characterization of Rhizobium japonicum hydrogen uptake genes. J Bacteriol. 1984 Sep;159(3):1006–1012. doi: 10.1128/jb.159.3.1006-1012.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapulnik Y., Phillips D. A. Sodium stimulation of uptake hydrogenase activity in symbiotic Rhizobium. Plant Physiol. 1986 Oct;82(2):494–498. doi: 10.1104/pp.82.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyser H. H., van Berkum P., Weber D. F. A Comparative Study of the Physiology of Symbioses Formed by Rhizobium japonicum with Glycine max, Vigna unguiculata, and Macroptilium atropurpurem. Plant Physiol. 1982 Dec;70(6):1626–1630. doi: 10.1104/pp.70.6.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyva A., Palacios J. M., Mozo T., Ruiz-Argüeso T. Cloning and characterization of hydrogen uptake genes from Rhizobium leguminosarum. J Bacteriol. 1987 Nov;169(11):4929–4934. doi: 10.1128/jb.169.11.4929-4934.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merberg D., O'Hara E. B., Maier R. J. Regulation of hydrogenase in Rhizobium japonicum: analysis of mutants altered in regulation by carbon substrates and oxygen. J Bacteriol. 1983 Dec;156(3):1236–1242. doi: 10.1128/jb.156.3.1236-1242.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo J., Villa A., Chamber M., Ruiz-Argüeso T. Occurrence of H(2)-Uptake Hydrogenases in Bradyrhizobium sp. (Lupinus) and Their Expression in Nodules of Lupinus spp. and Ornithopus compressus. Plant Physiol. 1989 Jan;89(1):78–85. doi: 10.1104/pp.89.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson L. M., Salminen S. O. Uptake hydrogenase activity and ATP formation in Rhizobium leguminosarum bacteroids. J Bacteriol. 1982 Aug;151(2):989–995. doi: 10.1128/jb.151.2.989-995.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brian M. R., Maier R. J. Molecular aspects of the energetics of nitrogen fixation in Rhizobium-legume symbioses. Biochim Biophys Acta. 1989 May 30;974(3):229–246. doi: 10.1016/s0005-2728(89)80239-7. [DOI] [PubMed] [Google Scholar]
- Sayavedra-Soto L. A., Powell G. K., Evans H. J., Morris R. O. Nucleotide sequence of the genetic loci encoding subunits of Bradyrhizobium japonicum uptake hydrogenase. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8395–8399. doi: 10.1073/pnas.85.22.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K. R., Evans H. J. Hydrogen evolution: A major factor affecting the efficiency of nitrogen fixation in nodulated symbionts. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1207–1211. doi: 10.1073/pnas.73.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]


