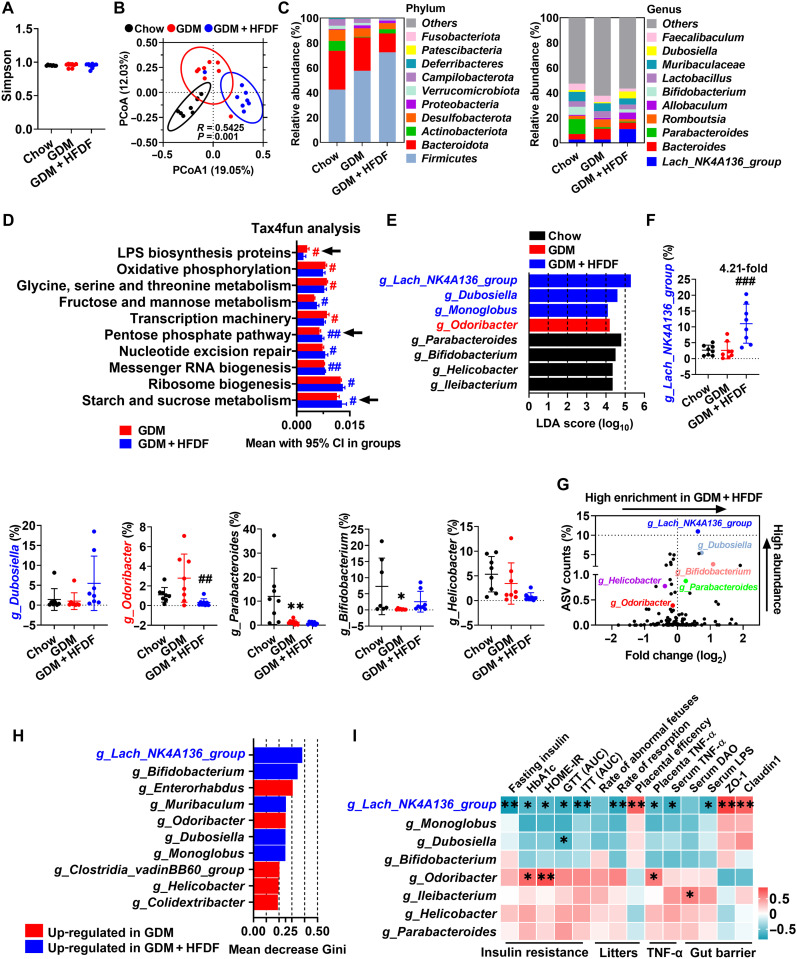
Fig. 6. Effects of HFDF on gut bacteria in GDM model.
Schematic of the maternal diet regimen and breeding is shown in Fig. 2A. At E12.5, fresh feces of mice were obtained, followed by 16S rRNA analysis (n = 8 mice per group). (A) Simpson index. (B) PCoA based on Bray-Curtis between Chow, GDM, and GDM + HFDF groups. (C) Average relative abundances of predominant taxa at phylum and genus level (n = 8 mice per group). (D) Relative abundance of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (top 10) in gut predicted by Tax4fun. (E) The most differentially abundant taxa between three groups identified by LDA effect size. Only LDA threshold of ≥4 taxa are shown. (F) Relative abundances of g_Lachnospiraceae_NK4A136_group, g_Dubosiella, g_Odoribacter, g_Parabacteroides, g_Bifidobacterium, and g_Helicobacter (n = 8 mice per group). g_Ileibacterium and g_Monoglobu are not shown because they were identified in only about half of the samples. (G) Volcano plot for the relative abundance distribution of microbial amplicon sequence variants (ASVs). Each symbol represents one bacterial taxon. (H) Identification of key bacterial genera by the random forest method using the 16S rRNA gene sequencing data of GDM and GDM + HFDF groups. Higher mean decrease Gini values indicate greater importance of the variable. (I) Pearson’s correlation coefficient analysis between the relative abundance of selected bacterial genera and insulin resistance, litters, TNF-α, or gut barrier parameters in GDM and GDM + HFDF mice. These parameters are based on the data from Figs. 2 to 5. Data were represented means ± SEM or means with 95% confidence interval (CI). P values were analyzed by one-way ANOVA followed by post hoc Tukey’s tests (A), unpaired Student’s t test (D), or one-way ANOVA followed by two-stage step-up false discovery rate (FDR) method of Benjamini, Krieger, and Yekutieli (F). * or #P < 0.05, ** or ##P < 0.01, and ###P < 0.001, relative to chow and GDM group, respectively.